RNA-Quast contents for Arabidopsis thaliana
S5.1: Short report for Arabidopsis thaliana assemblies
| METRICS/TRANSCRIPTS | Genes | Avg. number of exons per isoform | Transcripts | Transcripts > 500 bp | Transcripts > 1000 bp | Aligned | Uniquely aligned | Multiply aligned | Unaligned | Avg. aligned fraction | Avg. alignment length | Avg. mismatches per transcript | Misassemblies | Database coverage |
|---|
| Trinity | 32833 | 5.813 | 34617 | 21969 | 13357 | 34432 | 32388 | 44 | 185 | 0.993 | 978.213 | 1.315 | 1212 | 0.298 |
| Trans-ABySS | 32833 | 5.813 | 66004 | 31763 | 19714 | 65463 | 56460 | 176 | 541 | 0.979 | 745.088 | 0.612 | 7227 | 0.334 |
| Oases | 32833 | 5.813 | 57809 | 47279 | 32647 | 57441 | 48302 | 66 | 368 | 0.988 | 1232.688 | 3.471 | 6804 | 0.321 |
| SOAPdenovo-Trans | 32833 | 5.813 | 50876 | 17326 | 8709 | 50525 | 50289 | 119 | 351 | 0.997 | 561.746 | 0.099 | 51 | 0.299 |
| IDBA-Tran | 32833 | 5.813 | 32447 | 18147 | 9577 | 32366 | 32020 | 54 | 81 | 0.998 | 848.356 | 0.136 | 201 | 0.289 |
| Bridger | 32833 | 5.813 | 30017 | 20235 | 12882 | 29822 | 25907 | 45 | 195 | 0.99 | 990.349 | 2.427 | 1995 | 0.256 |
| BinPacker | 32833 | 5.813 | 5914 | 5458 | 3775 | 5891 | 3945 | 8 | 23 | 0.976 | 1312.171 | 5.902 | 1233 | 0.047 |
| Shannon | 32833 | 5.813 | 27685 | 18623 | 12018 | 27478 | 25912 | 37 | 207 | 0.996 | 1044.904 | 0.724 | 769 | 0.245 |
| rnaSPAdes | 32833 | 5.813 | 36953 | 18068 | 11160 | 36628 | 35821 | 91 | 325 | 0.995 | 807.422 | 0.341 | 505 | 0.3 |
| SPAdes
| 32833
| 5.813
| 29929
| 17390
| 11171
| 29691
| 27985
| 62
| 238
| 0.993
| 944.388
| 0.43
| 1189
| 0.279
|
| METRICS/TRANSCRIPTS | 50%-assembled genes | 95%-assembled genes | 50%-covered genes | 95%-covered genes | 50%-assembled isoforms | 95%-assembled isoforms | 50%-covered isoforms | 95%-covered isoforms | Mean isoform coverage | Mean isoform assembly | 50%-matched | 95%-matched | Unannotated | Mean fraction of transcript matched |
|---|
| Trinity | 12459 | 2196 | 14388 | 2542 | 13304 | 2302 | 15266 | 2651 | 0.684 | 0.625 | 30436 | 26763 | 887 | 0.92 |
| Trans-ABySS | 13755 | 1881 | 15660 | 2200 | 15283 | 1960 | 17334 | 2298 | 0.639 | 0.582 | 50342 | 41811 | 3066 | 0.866 |
| Oases | 13955 | 2599 | 14680 | 2812 | 15861 | 2753 | 16686 | 2981 | 0.734 | 0.707 | 46508 | 39238 | 788 | 0.933 |
| SOAPdenovo-Trans | 11457 | 1213 | 15357 | 1770 | 11470 | 1213 | 15395 | 1770 | 0.591 | 0.489 | 44555 | 42150 | 2479 | 0.887 |
| IDBA-Tran | 12672 | 1175 | 14963 | 1642 | 12694 | 1175 | 15017 | 1642 | 0.654 | 0.579 | 30039 | 28142 | 801 | 0.929 |
| Bridger | 11301 | 1861 | 12519 | 2076 | 11814 | 1909 | 13047 | 2127 | 0.666 | 0.623 | 24213 | 21017 | 876 | 0.911 |
| BinPacker | 2589 | 477 | 2609 | 488 | 2706 | 489 | 2729 | 500 | 0.751 | 0.745 | 3808 | 2929 | 23 | 0.917 |
| Shannon | 11168 | 1962 | 12207 | 2103 | 11606 | 2004 | 12658 | 2147 | 0.698 | 0.657 | 24594 | 22274 | 854 | 0.929 |
| rnaSPAdes | 12931 | 2233 | 15136 | 2607 | 13051 | 2237 | 15283 | 2613 | 0.672 | 0.603 | 32784 | 28995 | 1938 | 0.896 |
| SPAdes
| 12496
| 2318
| 13998
| 2570
| 12509
| 2318
| 14031
| 2570
| 0.681
| 0.632
| 25910
| 22414
| 1100
| 0.902
|
S5.2: Assembly completeness (sensitivity, true positive rate) for Arabidopsis thaliana assemblies
From the RNA-Quast manual page: "Assembly completeness (sensitivity). For the following metrics (calculated with reference genome and gene database) rnaQUAST attempts to select best-matching database isoforms for every transcript. Note that a single transcript can contribute to multiple isoforms in the case of, for example, paralogous genes or genomic repeats. At the same time, an isoform can be covered by multiple transcripts in the case of fragmented assembly or duplicated transcripts in the assembly." For further details see the RNA-Quast manual.
| METRICS/TRANSCRIPTS | Database coverage | Duplication ratio | Avg. number of transcripts mapped to one isoform | 50%-assembled genes | 95%-assembled genes | 50%-covered genes | 95%-covered genes | 50%-assembled isoforms | 95%-assembled isoforms | 50%-covered isoforms | 95%-covered isoforms | 50%-assembled exons | 95%-assembled exons | Mean isoform assembly | Mean isoform coverage | Mean exon coverage | Avg. percentage of isoform 50%-covered exons | Avg. percentage of isoform 95%-covered exons |
|---|
| Trinity | 0.298 | 1.128 | 1.517 | 12459 | 2196 | 14388 | 2542 | 13304 | 2302 | 15266 | 2651 | 91985 | 66950 | 0.625 | 0.684 | 0.893 | 0.736 | 0.42 |
| Trans-ABySS | 0.334 | 1.341 | 2.142 | 13755 | 1881 | 15660 | 2200 | 15283 | 1960 | 17334 | 2298 | 99841 | 71380 | 0.582 | 0.639 | 0.868 | 0.686 | 0.377 |
| Oases | 0.321 | 1.976 | 2.318 | 13955 | 2599 | 14680 | 2812 | 15861 | 2753 | 16686 | 2981 | 96063 | 71517 | 0.707 | 0.734 | 0.904 | 0.795 | 0.477 |
| SOAPdenovo-Trans | 0.299 | 1.008 | 1.963 | 11457 | 1213 | 15357 | 1770 | 11470 | 1213 | 15395 | 1770 | 94715 | 65284 | 0.489 | 0.591 | 0.858 | 0.63 | 0.329 |
| IDBA-Tran | 0.289 | 1.01 | 1.481 | 12672 | 1175 | 14963 | 1642 | 12694 | 1175 | 15017 | 1642 | 94428 | 68543 | 0.579 | 0.654 | 0.89 | 0.707 | 0.384 |
| Bridger | 0.256 | 1.067 | 1.372 | 11301 | 1861 | 12519 | 2076 | 11814 | 1909 | 13047 | 2127 | 81178 | 59493 | 0.623 | 0.666 | 0.887 | 0.715 | 0.407 |
| BinPacker | 0.047 | 1.149 | 1.203 | 2589 | 477 | 2609 | 488 | 2706 | 489 | 2729 | 500 | 15632 | 11737 | 0.745 | 0.751 | 0.911 | 0.818 | 0.489 |
| Shannon | 0.245 | 1.185 | 1.499 | 11168 | 1962 | 12207 | 2103 | 11606 | 2004 | 12658 | 2147 | 77697 | 57086 | 0.657 | 0.698 | 0.894 | 0.755 | 0.427 |
| rnaSPAdes | 0.3 | 1.027 | 1.608 | 12931 | 2233 | 15136 | 2607 | 13051 | 2237 | 15283 | 2613 | 95185 | 69477 | 0.603 | 0.672 | 0.889 | 0.721 | 0.404 |
| SPAdes | 0.279 | 1.007 | 1.406 | 12496 | 2318 | 13998 | 2570 | 12509 | 2318 | 14031 | 2570 | 89800 | 66775 | 0.632 | 0.681 | 0.895 | 0.733 | 0.419 |
S5.3: Assembly specificity (true negative rate) for Arabidopsis thaliana assemblies
From the RNA-Quast manual page: "Assembly specificity. To compute the following metrics we use only transcripts that have at least one significant alignment and are not misassembled." For further details see the RNA-Quast manual.
| METRICS/TRANSCRIPTS | Unannotated | 50%-matched | 95%-matched | Mean fraction of transcript matched | Mean fraction of block matched | 50%-matched blocks | 95%-matched blocks | Matched length | Unmatched length |
|---|
| Trinity | 887 | 30436 | 26763 | 0.92 | 0.961 | 0.963 | 0.94 | 30013045 | 1997896 |
| Trans-ABySS | 3066 | 50342 | 41811 | 0.866 | 0.96 | 0.961 | 0.938 | 39985550 | 2854902 |
| Oases | 788 | 46508 | 39238 | 0.933 | 0.956 | 0.959 | 0.937 | 56614558 | 3847059 |
| SOAPdenovo-Trans | 2479 | 44555 | 42150 | 0.887 | 0.966 | 0.963 | 0.95 | 26902613 | 1538113 |
| IDBA-Tran | 801 | 30039 | 28142 | 0.929 | 0.975 | 0.976 | 0.96 | 26093803 | 1200078 |
| Bridger | 876 | 24213 | 21017 | 0.911 | 0.962 | 0.964 | 0.943 | 24386684 | 1678489 |
| BinPacker | 23 | 3808 | 2929 | 0.917 | 0.942 | 0.945 | 0.918 | 4857682 | 495914 |
| Shannon | 854 | 24594 | 22274 | 0.929 | 0.971 | 0.973 | 0.955 | 25918151 | 1383207 |
| rnaSPAdes | 1938 | 32784 | 28995 | 0.896 | 0.968 | 0.972 | 0.944 | 27457743 | 1771614 |
| SPAdes | 1100 | 25910 | 22414 | 0.902 | 0.965 | 0.969 | 0.94 | 25038690 | 1699469 |
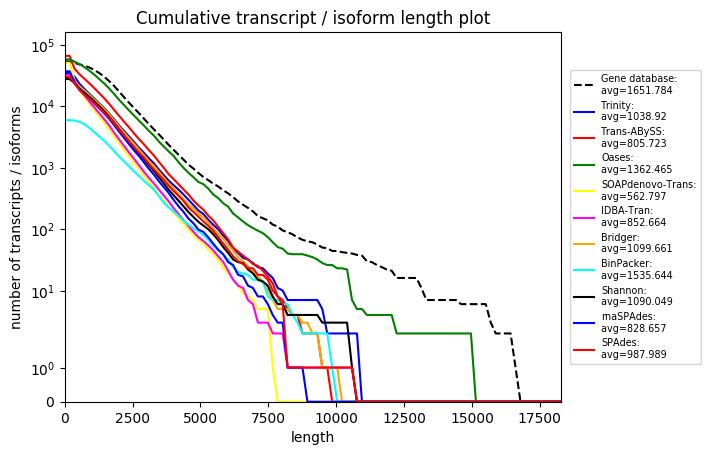
S5.4: Various comparison and single plots for Arabidopsis thaliana assemblies
Various comparison and single plots for Arabidopsis thaliana assemblies. For detailed descriptions of each plot please check the RNA-Quast manual page: