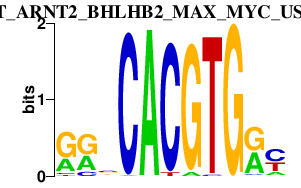
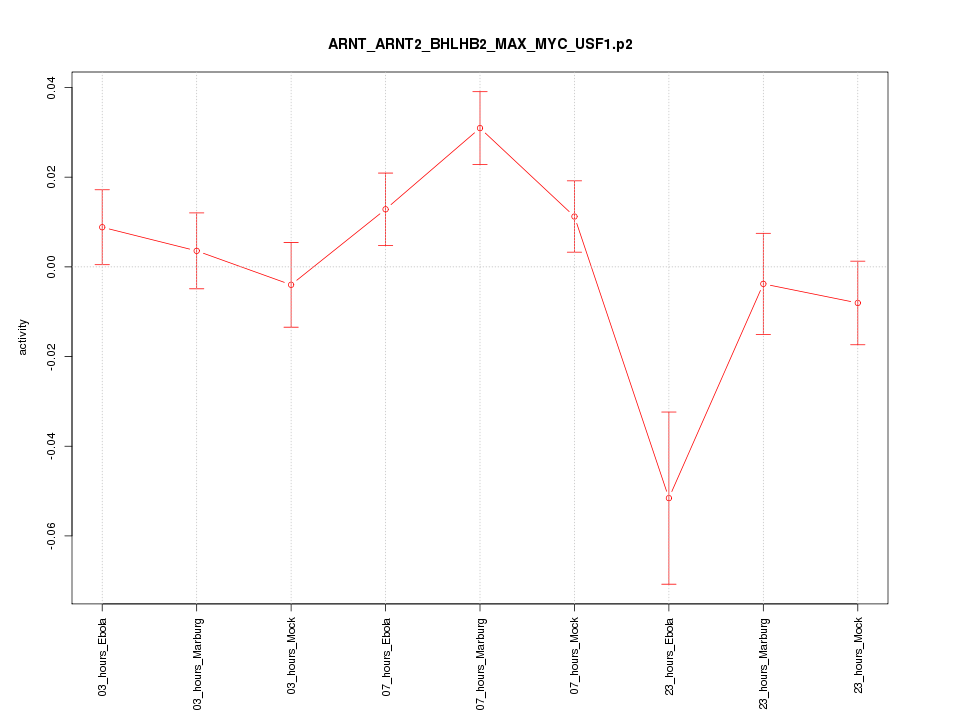
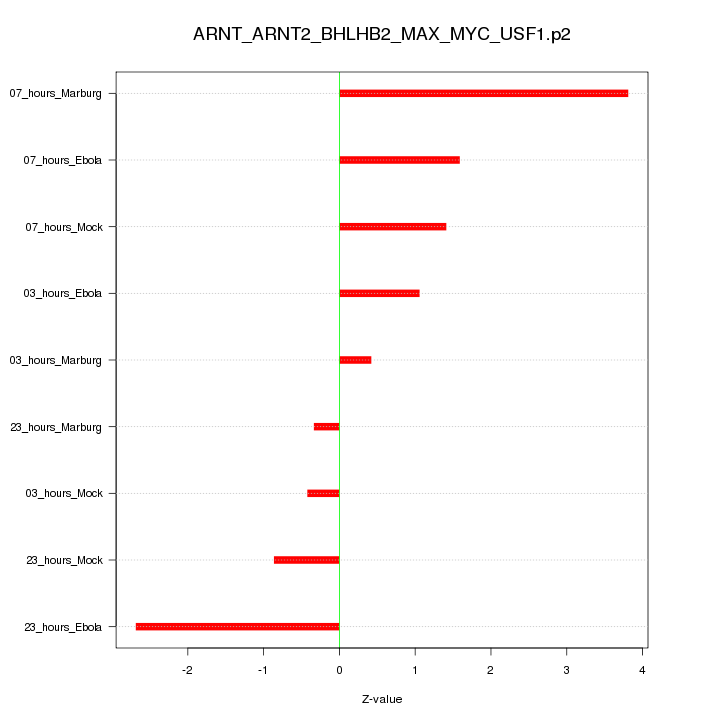
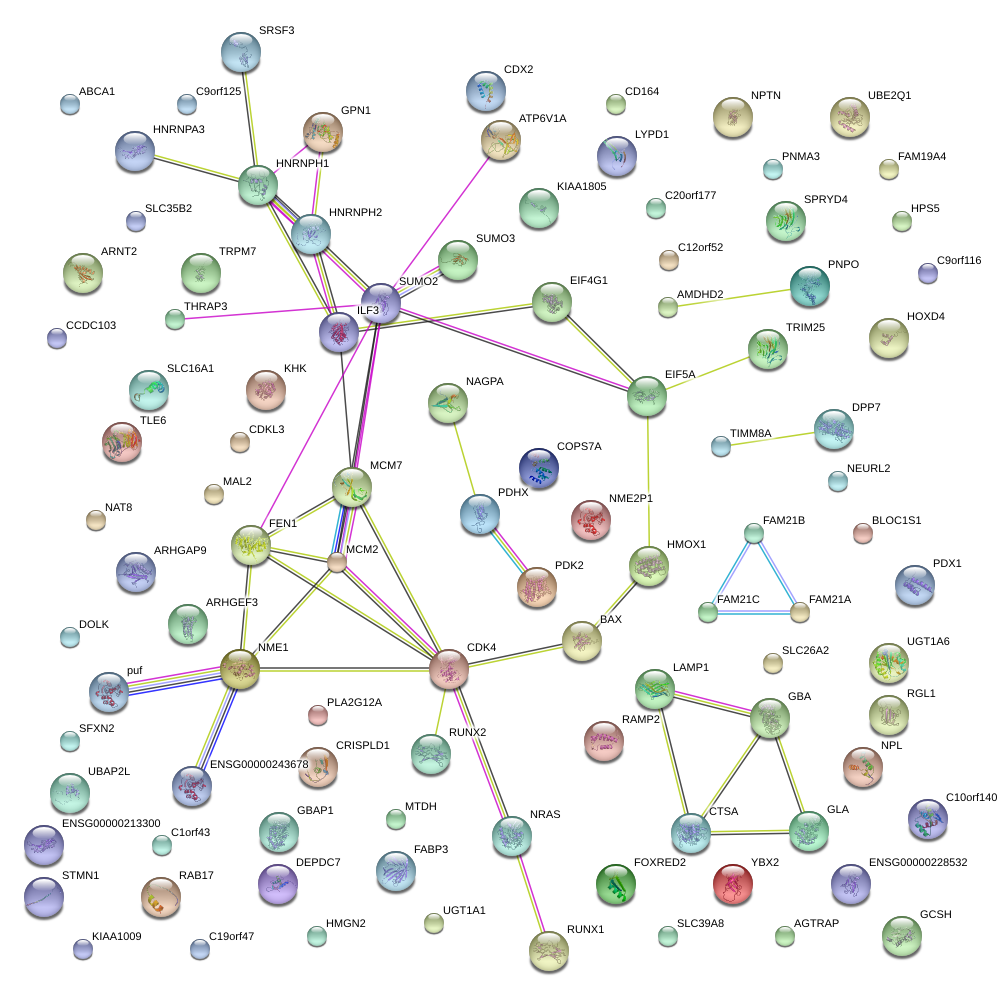
|
chr19_+_49458151
|
3.624
|
|
BAX
|
BCL2-associated X protein
|
|
chr17_+_46019022
|
3.451
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr6_-_84937313
|
3.246
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr20_+_44520021
|
3.049
|
|
CTSA
|
cathepsin A
|
|
chr12_+_112451229
|
3.011
|
|
|
|
|
chrX_-_100603673
|
3.003
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr9_-_131709783
|
2.943
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr10_+_104474297
|
2.861
|
NM_178858
|
SFXN2
|
sideroflexin 2
|
|
chr1_-_155210887
|
2.741
|
|
GBA
|
glucosidase, beta, acid
|
|
chr1_-_155211049
|
2.572
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr16_+_2570322
|
2.535
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr20_+_44519924
|
2.437
|
|
CTSA
|
cathepsin A
|
|
chr19_+_49458129
|
2.396
|
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_154531083
|
2.377
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr20_+_44519965
|
2.355
|
|
CTSA
|
cathepsin A
|
|
chr19_+_49458031
|
2.318
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chrX_+_100663213
|
2.256
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_+_11796211
|
2.187
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr1_+_183605207
|
2.168
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr12_+_56862300
|
2.148
|
NM_207344
|
SPRYD4
|
SPRY domain containing 4
|
|
chr1_+_154193294
|
2.148
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chrX_+_100663205
|
2.147
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr8_+_75896707
|
2.108
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr13_+_28494136
|
2.036
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr5_+_149340346
|
2.027
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chrX_+_100663229
|
2.003
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr20_+_58515408
|
1.984
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr17_+_40913233
|
1.926
|
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr3_-_57113292
|
1.914
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr2_+_177016112
|
1.897
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr9_-_107690526
|
1.896
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr3_+_113465901
|
1.865
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr17_+_46018888
|
1.853
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr9_-_107690435
|
1.823
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr17_+_40913211
|
1.756
|
NM_005854
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr4_-_110650965
|
1.753
|
NM_030821
|
PLA2G12A
|
phospholipase A2, group XIIA
|
|
chr17_-_54991399
|
1.740
|
NM_005082
|
TRIM25
|
tripartite motif containing 25
|
|
chr9_-_138391696
|
1.717
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr2_+_27309610
|
1.709
|
NM_000221
NM_006488
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr9_-_107690336
|
1.704
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_-_31845818
|
1.697
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr1_+_36689996
|
1.674
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr19_+_10764987
|
1.615
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr22_-_36903033
|
1.597
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr1_+_26798961
|
1.585
|
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr16_-_5083933
|
1.559
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr3_+_113465860
|
1.547
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr19_-_40854410
|
1.530
|
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr13_+_113951537
|
1.528
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr5_+_149340437
|
1.523
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr15_-_73925658
|
1.522
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr9_-_140009167
|
1.511
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr21_-_46237974
|
1.505
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr15_-_73925555
|
1.503
|
|
NPTN
|
neuroplastin
|
|
chr22_+_35777087
|
1.494
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr20_+_44520203
|
1.486
|
|
CTSA
|
cathepsin A
|
|
chr17_+_42977141
|
1.480
|
NM_213607
|
CCDC103
|
coiled-coil domain containing 103
|
|
chr21_-_36421586
|
1.480
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr21_-_36421630
|
1.473
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_26798901
|
1.452
|
NM_005517
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr21_-_46237978
|
1.448
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr21_-_36421578
|
1.447
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr2_+_178077421
|
1.432
|
NM_194247
|
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chr16_-_81129882
|
1.426
|
NM_004483
|
GCSH
|
glycine cleavage system protein H (aminomethyl carrier)
|
|
chr20_-_44519629
|
1.423
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr2_-_238499317
|
1.423
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr17_+_79885567
|
1.401
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr3_+_113465910
|
1.399
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr8_+_98656428
|
1.395
|
|
MTDH
|
metadherin
|
|
chr5_+_149340287
|
1.387
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr2_-_133427624
|
1.385
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr1_-_26232890
|
1.364
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr22_-_36903086
|
1.364
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr19_+_10764896
|
1.363
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr1_-_154193073
|
1.363
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chrX_-_100662788
|
1.356
|
|
GLA
|
galactosidase, alpha
|
|
chr12_-_58146108
|
1.356
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr16_-_5083913
|
1.355
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr1_-_115259381
|
1.349
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr3_-_68981674
|
1.341
|
NM_001005527
NM_182522
|
FAM19A4
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4
|
|
chr11_-_71814268
|
1.331
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr19_+_10765106
|
1.320
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr2_+_27805892
|
1.305
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr11_+_33037397
|
1.304
|
NM_001077242
|
DEPDC7
|
DEP domain containing 7
|
|
chr21_-_46237966
|
1.293
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr1_+_11796128
|
1.293
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr9_-_104249333
|
1.288
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr12_+_56109851
|
1.275
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr12_-_58146110
|
1.266
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chrX_-_100662905
|
1.266
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr3_+_127317274
|
1.264
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr8_+_98656494
|
1.261
|
|
MTDH
|
metadherin
|
|
chrX_-_100603956
|
1.251
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr2_+_234668914
|
1.247
|
NM_000463
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr19_-_40854283
|
1.238
|
NM_178830
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr11_-_71814335
|
1.232
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chrX_+_152224862
|
1.231
|
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr8_+_120220554
|
1.224
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr11_+_61560108
|
1.220
|
NM_004111
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr12_+_113623364
|
1.215
|
NM_032848
|
C12orf52
|
chromosome 12 open reading frame 52
|
|
chr3_+_127317271
|
1.212
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr13_-_28543143
|
1.201
|
|
CDX2
|
caudal type homeobox 2
|
|
chr3_+_184032319
|
1.190
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr13_-_51417831
|
1.185
|
NM_198989
|
DLEU7
|
deleted in lymphocytic leukemia, 7
|
|
chr10_-_21814610
|
1.183
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr17_-_7197896
|
1.181
|
|
YBX2
|
Y box binding protein 2
|
|
chr17_+_48172508
|
1.179
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr6_+_36562135
|
1.169
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr17_+_49244082
|
1.167
|
NM_001018138
|
NME2
|
non-metastatic cells 2, protein (NM23B) expressed in
|
|
chr11_-_71814294
|
1.157
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr15_-_50978921
|
1.153
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr6_-_109703661
|
1.139
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr10_+_51827710
|
1.137
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr17_+_7211693
|
1.136
|
NM_001143762
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr19_+_2977480
|
1.136
|
NM_001143986
NM_024760
|
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila)
|
|
chr1_+_182758871
|
1.132
|
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr20_+_44519590
|
1.128
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr1_-_113498828
|
1.118
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr17_+_7211260
|
1.117
|
NM_001143761
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr4_-_103266577
|
1.109
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr11_-_18343670
|
1.109
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr12_+_6833425
|
1.103
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr6_-_44225024
|
1.102
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr5_-_133706717
|
1.099
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr16_-_74401991
|
1.098
|
|
LOC283922
|
pyruvate dehydrogenase phosphatase regulatory subunit pseudogene
|
|
chr19_+_45909466
|
1.097
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr13_+_49684388
|
1.096
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr3_+_127317212
|
1.087
|
NM_004526
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr10_+_99344421
|
1.079
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chrX_+_100663120
|
1.074
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr17_+_48450579
|
1.071
|
NM_001166131
NM_152463
|
EME1
|
essential meiotic endonuclease 1 homolog 1 (S. pombe)
|
|
chr20_-_58515258
|
1.067
|
NM_006242
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D
|
|
chr1_+_11796177
|
1.066
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr1_-_154193044
|
1.056
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chrX_+_152224765
|
1.051
|
NM_013364
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr12_+_56109832
|
1.048
|
|
BLOC1S1-RDH5
BLOC1S1
|
BLOC1S1-RDH5 readthrough
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr10_+_25305507
|
1.046
|
NM_024838
|
THNSL1
|
threonine synthase-like 1 (S. cerevisiae)
|
|
chr14_+_20923341
|
1.045
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr1_-_113498615
|
1.042
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr15_+_45315301
|
1.038
|
NM_003104
|
SORD
|
sorbitol dehydrogenase
|
|
chr6_-_109804432
|
1.038
|
NM_001164313
NM_014797
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr2_+_85766100
|
1.036
|
NM_005911
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr1_-_204329043
|
1.033
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr11_-_71814371
|
1.032
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr13_+_42846255
|
1.031
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr5_-_111312522
|
1.029
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_6633531
|
1.028
|
|
SRD5A1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1)
|
|
chr12_-_58146025
|
1.027
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_+_112451261
|
1.026
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr5_-_180288275
|
1.021
|
NM_001172638
NM_152283
|
ZFP62
|
zinc finger protein 62 homolog (mouse)
|
|
chr16_-_4897265
|
1.018
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr1_-_26232643
|
1.015
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr20_+_18118473
|
1.015
|
NM_001164811
|
PET117
|
cytochrome c oxidase assembly factor-like
|
|
chr8_+_98656329
|
1.008
|
NM_178812
|
MTDH
|
metadherin
|
|
chr4_-_100871431
|
1.003
|
|
H2AFZ
|
H2A histone family, member Z
|
|
chr12_+_6833149
|
0.997
|
NM_001164094
NM_016319
NM_001164095
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr17_+_55163077
|
0.996
|
NM_001242903
NM_001242902
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr6_+_36562089
|
0.994
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr21_-_36421195
|
0.991
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr17_-_7197811
|
0.985
|
|
YBX2
|
Y box binding protein 2
|
|
chrX_-_102510048
|
0.983
|
NM_001006684
NM_153333
|
TCEAL8
|
transcription elongation factor A (SII)-like 8
|
|
chr22_+_35777051
|
0.978
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr12_+_117176085
|
0.976
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr11_+_61560401
|
0.974
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr10_-_72141347
|
0.968
|
NM_207119
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr3_+_184032414
|
0.967
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr7_-_99774821
|
0.964
|
NM_152742
|
GPC2
|
glypican 2
|
|
chr1_-_154600446
|
0.954
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr14_+_77564570
|
0.953
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr6_+_151773421
|
0.952
|
NM_024573
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr12_-_58146118
|
0.948
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr2_-_25474948
|
0.948
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr4_-_25032306
|
0.947
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr19_+_33793743
|
0.945
|
|
LOC80054
|
uncharacterized LOC80054
|
|
chrX_+_149737019
|
0.944
|
NM_000252
|
MTM1
|
myotubularin 1
|
|
chr17_+_42422490
|
0.938
|
NM_002087
|
GRN
|
granulin
|
|
chr16_-_74401792
|
0.923
|
|
|
|
|
chr11_+_61560352
|
0.921
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr12_-_65153148
|
0.919
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr9_-_37904098
|
0.918
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr1_+_226016428
|
0.912
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr1_-_23670743
|
0.910
|
NM_001102397
NM_001102398
NM_001102399
NM_005826
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr14_-_47120940
|
0.908
|
NM_080746
|
RPL10L
|
ribosomal protein L10-like
|
|
chr11_+_12132063
|
0.908
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr1_-_183604885
|
0.903
|
NM_005717
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr16_+_22217369
|
0.898
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr13_+_113951589
|
0.892
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chrX_+_149737083
|
0.892
|
|
MTM1
|
myotubularin 1
|
|
chr1_-_111743261
|
0.891
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr12_-_65153182
|
0.890
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr2_+_173292344
|
0.889
|
|
ITGA6
|
integrin, alpha 6
|
|
chr10_-_46090191
|
0.887
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr5_+_126113050
|
0.884
|
|
LMNB1
|
lamin B1
|
|
chr17_-_18266691
|
0.881
|
NM_004169
NM_148918
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble)
|
|
chr8_+_98788014
|
0.878
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr12_-_49318663
|
0.870
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr20_-_39766522
|
0.868
|
|
|
|
|
chr1_-_166845533
|
0.867
|
NM_053053
|
TADA1
|
transcriptional adaptor 1
|
|
chr3_-_197686837
|
0.867
|
NM_032263
|
IQCG
|
IQ motif containing G
|
|
chr5_+_126112847
|
0.865
|
|
LMNB1
|
lamin B1
|