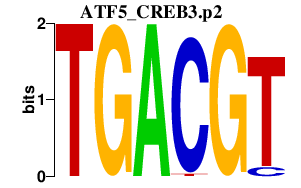
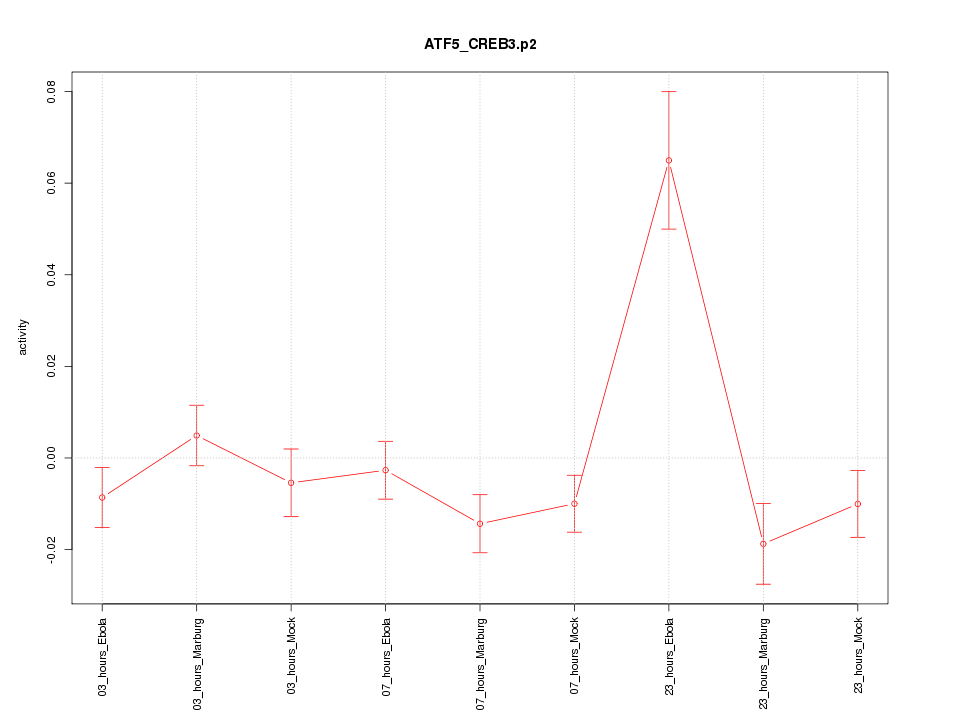
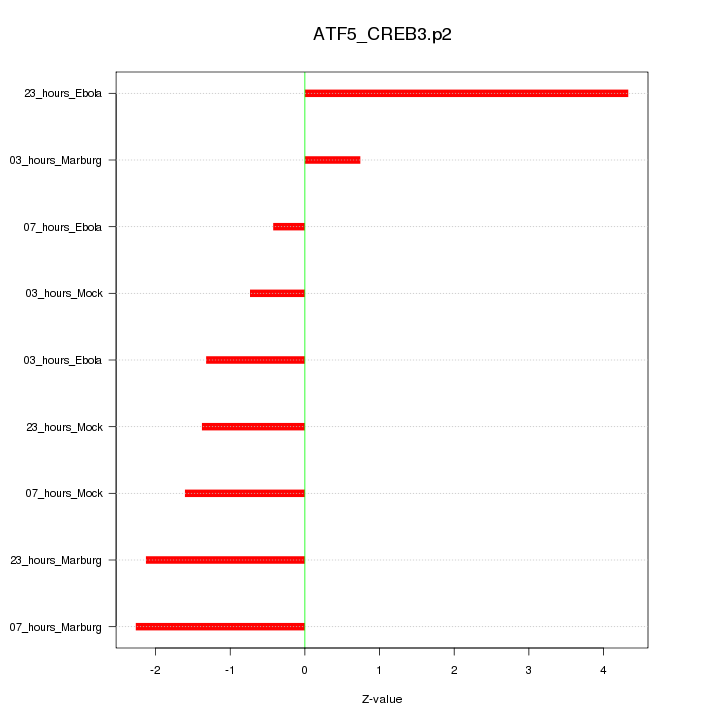
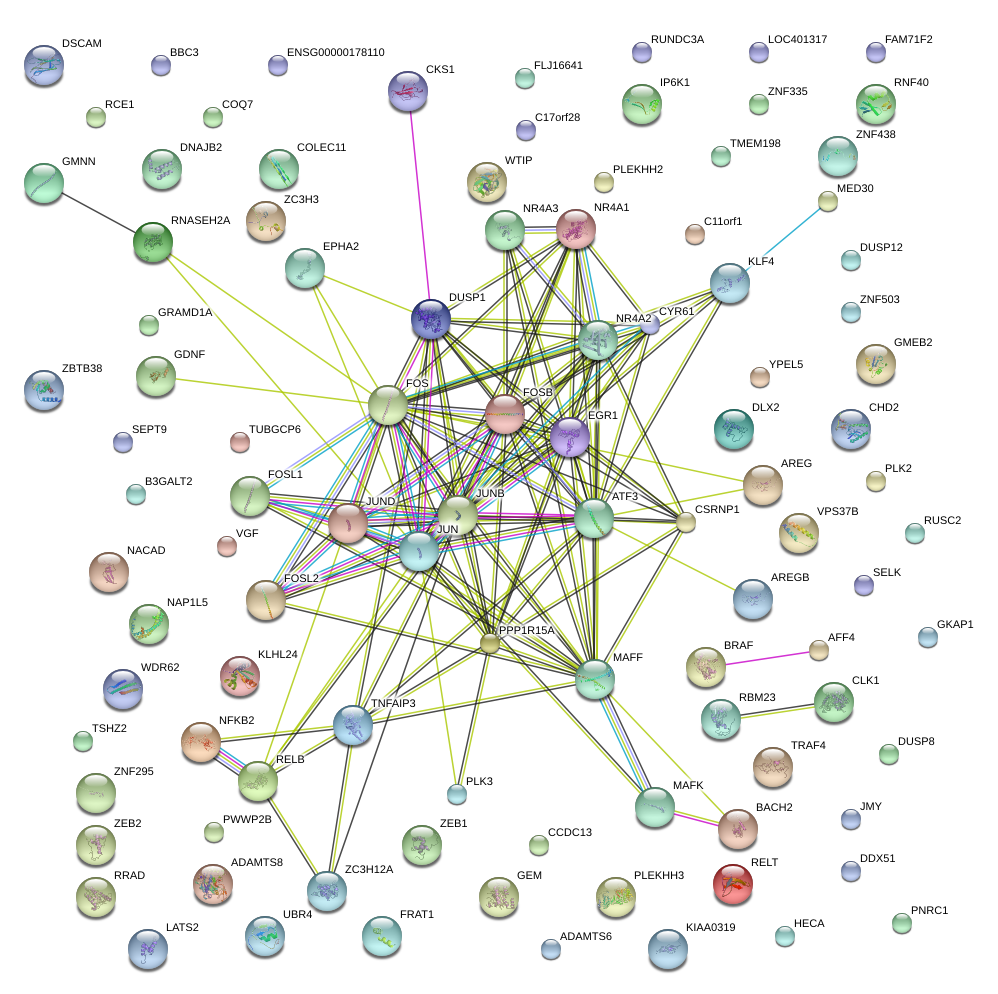
|
chr19_+_45971249
|
10.103
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr12_+_52445185
|
7.873
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr14_+_75745480
|
7.124
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr11_-_1593149
|
6.269
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr19_+_49375679
|
6.061
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr19_+_49375677
|
6.061
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr4_+_75480628
|
5.961
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr9_+_102584136
|
5.943
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr19_+_49375638
|
5.896
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr7_+_28448970
|
4.500
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr8_-_95274540
|
4.461
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr6_+_89791510
|
4.363
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr5_-_172198160
|
4.317
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr6_-_91006580
|
4.312
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr5_+_137801178
|
4.277
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr2_+_28615771
|
4.258
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr22_+_38597938
|
4.237
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr6_-_91006460
|
4.111
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_+_28615695
|
4.110
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr8_-_95274520
|
3.970
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr5_+_137801166
|
3.787
|
|
EGR1
|
early growth response 1
|
|
chr7_-_100808832
|
3.679
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr22_+_38598065
|
3.628
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr12_-_123380608
|
3.568
|
|
|
|
|
chr1_+_212781969
|
3.432
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr1_+_212782103
|
3.356
|
|
ATF3
|
activating transcription factor 3
|
|
chr2_+_28615668
|
3.344
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr10_+_104154228
|
3.241
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr21_-_43430448
|
2.920
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr16_-_66959418
|
2.887
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr8_+_128806720
|
2.779
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr4_-_89618928
|
2.662
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr12_-_123380682
|
2.599
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr3_-_8543327
|
2.490
|
|
LOC100288428
|
uncharacterized LOC100288428
|
|
chr20_-_44600939
|
2.483
|
|
ZNF335
|
zinc finger protein 335
|
|
chr3_-_42814734
|
2.442
|
NM_144719
|
CCDC13
|
coiled-coil domain containing 13
|
|
chr19_-_18392048
|
2.409
|
|
JUND
|
jun D proto-oncogene
|
|
chr12_-_123380628
|
2.397
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr7_+_1577118
|
2.387
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr1_+_86046434
|
2.378
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr11_-_65667809
|
2.372
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr17_-_40828078
|
2.345
|
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr2_+_220408352
|
2.289
|
|
TMEM198
|
transmembrane protein 198
|
|
chr3_+_50297644
|
2.259
|
|
|
|
|
chr19_+_45504686
|
2.241
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr14_-_23388294
|
2.231
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr11_+_73087396
|
2.218
|
NM_152222
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr19_-_18392413
|
2.214
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr10_-_31320772
|
2.178
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr2_+_220408742
|
2.162
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr2_-_145277568
|
2.135
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr13_-_21635629
|
2.133
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr2_-_157189040
|
2.129
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr2_-_172967627
|
2.120
|
|
DLX2
|
distal-less homeobox 2
|
|
chr14_-_23388380
|
2.112
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr19_+_35491225
|
2.081
|
NM_001136199
NM_020895
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr13_-_21635703
|
2.060
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr19_-_47734215
|
2.053
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr15_+_93447702
|
2.038
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr5_-_57755787
|
2.027
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr19_+_12902295
|
2.005
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr1_+_37940074
|
1.991
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr20_-_44600787
|
1.972
|
NM_022095
|
ZNF335
|
zinc finger protein 335
|
|
chr11_-_65667860
|
1.958
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr12_-_132628841
|
1.952
|
NM_175066
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr1_-_193155723
|
1.926
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr5_-_37839781
|
1.909
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr2_-_201729351
|
1.869
|
|
CLK1
|
CDC-like kinase 1
|
|
chr11_+_66610882
|
1.862
|
NM_001032279
NM_005133
|
RCE1
|
RCE1 homolog, prenyl protein protease (S. cerevisiae)
|
|
chr20_-_62258422
|
1.860
|
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr6_+_135818938
|
1.857
|
|
LINC00271
|
long intergenic non-protein coding RNA 271
|
|
chr1_-_19536741
|
1.840
|
NM_020765
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4
|
|
chr2_-_201729241
|
1.836
|
NM_001162407
|
CLK1
|
CDC-like kinase 1
|
|
chr3_-_53925871
|
1.812
|
NM_021237
|
SELK
|
selenoprotein K
|
|
chr1_-_16482426
|
1.809
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr19_-_18392436
|
1.806
|
|
JUND
|
jun D proto-oncogene
|
|
chr11_+_111749947
|
1.793
|
NM_022761
|
C11orf1
|
chromosome 11 open reading frame 1
|
|
chr1_+_45266035
|
1.791
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr2_-_145277879
|
1.787
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr8_-_144623579
|
1.782
|
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr10_+_104155431
|
1.743
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr7_+_1580009
|
1.718
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr17_-_72968790
|
1.713
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr5_+_78531848
|
1.706
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr1_-_713985
|
1.704
|
|
LOC100288069
|
general transcription factor IIi pseudogene
|
|
chr10_+_134210691
|
1.697
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr7_-_140624280
|
1.695
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr6_+_139456240
|
1.667
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr2_+_220144024
|
1.653
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr10_-_77161424
|
1.646
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr3_+_141087298
|
1.645
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr1_+_45266048
|
1.624
|
|
PLK3
|
polo-like kinase 3
|
|
chr20_-_62258330
|
1.603
|
NM_012384
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr9_-_86432546
|
1.600
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr1_-_59249687
|
1.600
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr19_+_12902286
|
1.581
|
|
|
|
|
chr3_-_39195101
|
1.567
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr2_+_30369749
|
1.565
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr2_-_201729377
|
1.560
|
NM_004071
|
CLK1
|
CDC-like kinase 1
|
|
chr17_-_72968750
|
1.552
|
|
|
|
|
chr17_+_27071008
|
1.534
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr11_+_73087670
|
1.529
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr5_-_64777703
|
1.514
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr17_+_42385780
|
1.510
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr3_+_183353355
|
1.509
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr10_-_77161539
|
1.507
|
|
ZNF503
|
zinc finger protein 503
|
|
chr2_+_43864405
|
1.496
|
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr3_-_39195034
|
1.481
|
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr5_+_78532338
|
1.471
|
|
|
|
|
chr3_+_156544075
|
1.468
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr7_-_45128492
|
1.454
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr6_+_138188352
|
1.452
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr5_-_132299248
|
1.434
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr8_+_118532970
|
1.434
|
|
MED30
|
mediator complex subunit 30
|
|
chr2_+_43864433
|
1.427
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr9_+_35538890
|
1.425
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_128312345
|
1.399
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr16_+_30773619
|
1.399
|
|
RNF40
|
ring finger protein 40
|
|
chr22_-_50683372
|
1.391
|
NM_020461
|
TUBGCP6
|
tubulin, gamma complex associated protein 6
|
|
chr9_-_110252045
|
1.385
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr3_-_49823626
|
1.365
|
NM_001242829
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr10_+_99079017
|
1.359
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr9_+_35538628
|
1.356
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr9_-_110251754
|
1.354
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr5_-_132299263
|
1.344
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr19_+_34972866
|
1.330
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr20_+_51589752
|
1.320
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr11_-_67141008
|
1.317
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr19_+_35491329
|
1.316
|
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr4_-_175204617
|
1.315
|
|
FBXO8
|
F-box protein 8
|
|
chr19_-_42721802
|
1.311
|
NM_133328
|
DEDD2
|
death effector domain containing 2
|
|
chr3_-_49823953
|
1.303
|
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr11_-_66610703
|
1.280
|
|
|
|
|
chr3_+_150125983
|
1.264
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr1_+_45265896
|
1.261
|
|
PLK3
|
polo-like kinase 3
|
|
chr8_-_11324184
|
1.259
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr2_+_10184371
|
1.257
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr4_+_668250
|
1.256
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr4_-_926081
|
1.256
|
|
GAK
|
cyclin G associated kinase
|
|
chr1_-_32110464
|
1.245
|
|
PEF1
|
penta-EF-hand domain containing 1
|
|
chr3_-_49823972
|
1.239
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr6_+_30295007
|
1.237
|
NM_172016
|
TRIM39
|
tripartite motif containing 39
|
|
chr4_-_926101
|
1.219
|
|
GAK
|
cyclin G associated kinase
|
|
chr19_-_3971032
|
1.203
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr13_+_73633097
|
1.203
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr16_+_54965110
|
1.202
|
NM_001252197
NM_005853
|
IRX5
|
iroquois homeobox 5
|
|
chr11_-_3818706
|
1.192
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr5_-_132299191
|
1.192
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr2_+_33172368
|
1.173
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr19_-_3971089
|
1.170
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr18_+_23806385
|
1.167
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chrX_+_49126299
|
1.165
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chrX_+_49126326
|
1.158
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr2_+_30369800
|
1.158
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr1_+_157963310
|
1.153
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr17_-_17399494
|
1.149
|
NM_001199989
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr19_+_55987698
|
1.147
|
NM_033113
|
ZNF628
|
zinc finger protein 628
|
|
chr8_-_29207795
|
1.146
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr15_+_90544608
|
1.142
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chrX_+_49126480
|
1.139
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr15_+_78556485
|
1.134
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr1_-_214724974
|
1.121
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr11_-_65667858
|
1.120
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr16_+_18995508
|
1.118
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr22_+_42229105
|
1.117
|
NM_004599
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr16_+_18995255
|
1.111
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr17_-_8151373
|
1.110
|
NM_025099
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr4_-_54930466
|
1.108
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr3_+_42977833
|
1.103
|
NM_001205272
|
KRBOX1
|
KRAB box domain containing 1
|
|
chr17_-_8151353
|
1.103
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr11_+_64692052
|
1.102
|
NM_006244
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta
|
|
chr17_-_76836852
|
1.101
|
NM_025090
|
USP36
|
ubiquitin specific peptidase 36
|
|
chr2_+_220071493
|
1.099
|
NM_138802
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr4_-_69215699
|
1.096
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr22_-_50682915
|
1.090
|
|
TUBGCP6
|
tubulin, gamma complex associated protein 6
|
|
chr17_-_76836718
|
1.089
|
|
USP36
|
ubiquitin specific peptidase 36
|
|
chr11_+_66059364
|
1.087
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr3_+_50306305
|
1.075
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr4_-_2010702
|
1.074
|
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr13_+_45915479
|
1.069
|
|
LOC100190939
|
uncharacterized LOC100190939
|
|
chr16_-_54320367
|
1.065
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr2_+_70142236
|
1.063
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr7_+_123295860
|
1.059
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr11_-_116968986
|
1.054
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr21_-_44846927
|
1.052
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr4_-_926114
|
1.049
|
|
GAK
|
cyclin G associated kinase
|
|
chrX_-_52987610
|
1.047
|
NM_001242491
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr15_+_83478329
|
1.046
|
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chr16_-_67450149
|
1.043
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr6_-_70506472
|
1.037
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr15_-_40401041
|
1.034
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr6_+_43445255
|
1.032
|
NM_001146016
NM_001146018
NM_001146019
NM_001146020
NM_080604
|
TJAP1
|
tight junction associated protein 1 (peripheral)
|
|
chr2_-_157186681
|
1.025
|
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr6_+_43445304
|
1.020
|
|
TJAP1
|
tight junction associated protein 1 (peripheral)
|
|
chr19_+_39390570
|
1.013
|
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr5_-_175964824
|
1.010
|
|
RNF44
|
ring finger protein 44
|
|
chr4_+_103422553
|
1.007
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr1_-_214724844
|
1.007
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr2_+_169659099
|
1.006
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr14_+_105331581
|
0.998
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|