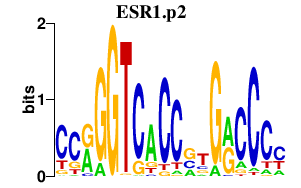
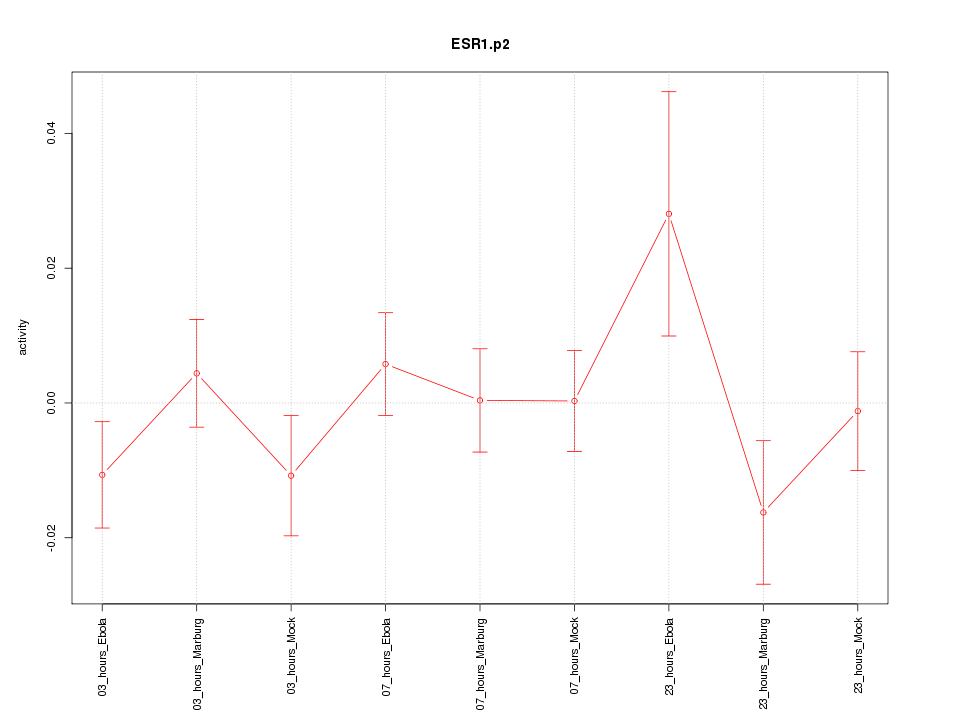
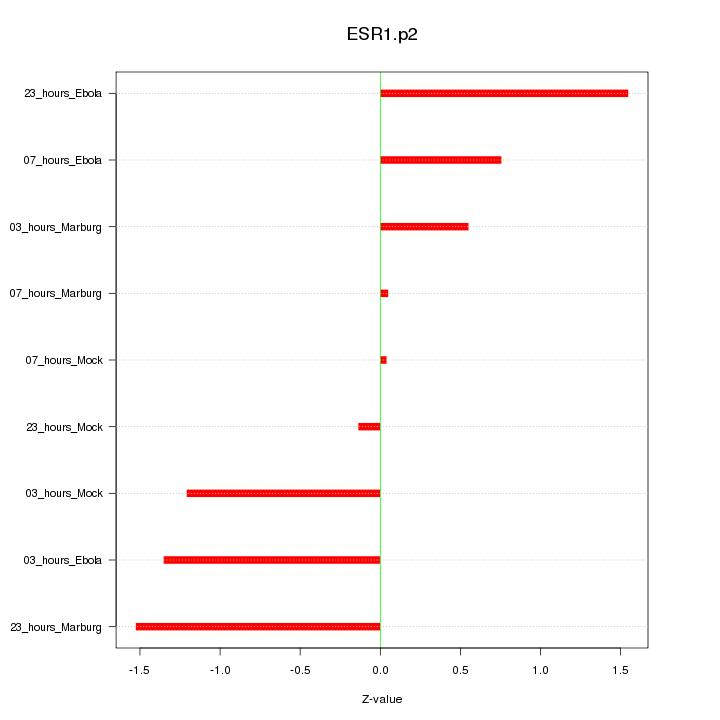
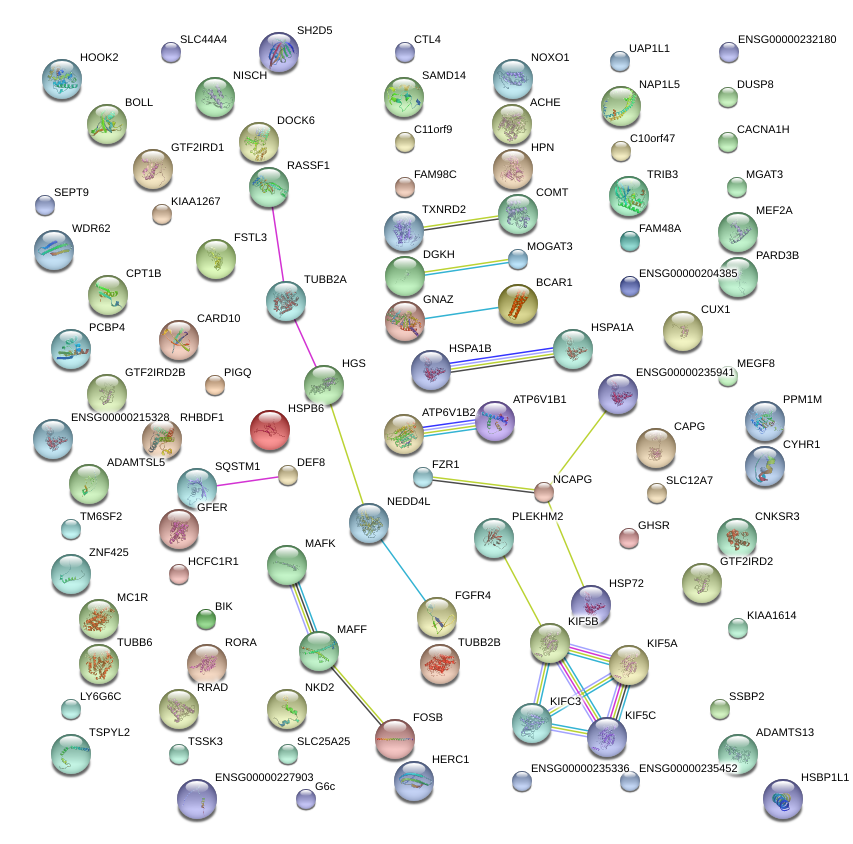
|
chr18_+_77724575
|
1.625
|
NM_001136180
|
HSBP1L1
|
heat shock factor binding protein 1-like 1
|
|
chr16_-_75285383
|
1.047
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr15_-_64126112
|
0.966
|
|
HERC1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1
|
|
chr16_+_1203240
|
0.958
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr22_-_19929447
|
0.925
|
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr7_+_1570321
|
0.907
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr19_+_676346
|
0.873
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr19_+_38893766
|
0.840
|
NM_174905
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr22_+_19159198
|
0.788
|
|
LOC100652736
|
uncharacterized LOC100652736
|
|
chr19_+_45971249
|
0.743
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr1_+_16010838
|
0.717
|
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr7_+_1577118
|
0.707
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr2_-_198650008
|
0.655
|
NM_033030
|
BOLL
|
bol, boule-like (Drosophila)
|
|
chr16_+_1203737
|
0.655
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr7_-_65235688
|
0.636
|
|
LOC441242
|
uncharacterized LOC441242
|
|
chr16_+_89985268
|
0.633
|
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor)
|
|
chr8_+_128806720
|
0.632
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr22_-_19929333
|
0.625
|
NM_006440
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr9_+_139972195
|
0.622
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr16_-_122573
|
0.608
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr11_-_1593149
|
0.599
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr16_+_90015827
|
0.580
|
NM_001242819
|
DEF8
|
differentially expressed in FDCP 8 homolog (mouse)
|
|
chr19_+_3522953
|
0.572
|
NM_001136197
NM_001136198
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr19_+_3506264
|
0.568
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr16_-_3074225
|
0.567
|
NM_001002017
NM_001002018
NM_017885
|
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent)
|
|
chr6_-_31689455
|
0.566
|
NM_025261
|
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr22_-_19929250
|
0.561
|
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr18_+_55711890
|
0.556
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_+_180882312
|
0.551
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr20_+_361938
|
0.551
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr3_-_50374749
|
0.545
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr22_+_43506753
|
0.540
|
NM_001197
|
BIK
|
BCL2-interacting killer (apoptosis-inducing)
|
|
chr2_+_149633173
|
0.531
|
|
|
|
|
chr7_-_100493481
|
0.528
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr4_-_89618928
|
0.524
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr3_-_50374875
|
0.523
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr19_+_7660698
|
0.522
|
NM_001080429
NM_020902
|
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3
|
|
chr22_-_19929322
|
0.516
|
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr6_+_31783280
|
0.515
|
NM_005345
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr19_-_1513187
|
0.514
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr3_-_50374795
|
0.512
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr16_+_619969
|
0.512
|
NM_148920
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q
|
|
chr16_-_2031359
|
0.512
|
NM_144603
|
NOXO1
|
NADPH oxidase organizer 1
|
|
chr19_-_19384022
|
0.505
|
NM_001001524
|
TM6SF2
|
transmembrane 6 superfamily member 2
|
|
chr6_+_31783314
|
0.505
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr22_-_37915209
|
0.503
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr15_-_64126081
|
0.502
|
|
HERC1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1
|
|
chr5_-_1112106
|
0.499
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr17_+_79651035
|
0.499
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr17_+_79650961
|
0.496
|
NM_004712
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr22_+_23412364
|
0.492
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr7_+_101460881
|
0.491
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr22_+_38597938
|
0.491
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr19_+_35532640
|
0.490
|
|
HPN
|
hepsin
|
|
chr9_+_136287443
|
0.475
|
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr17_+_79651052
|
0.472
|
|
|
|
|
chr15_+_100106179
|
0.472
|
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr8_-_145690417
|
0.471
|
NM_138496
|
CYHR1
|
cysteine/histidine-rich 1
|
|
chr17_-_44302716
|
0.470
|
NM_001193465
|
KIAA1267
|
KIAA1267
|
|
chr19_+_38893804
|
0.464
|
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr2_+_71162995
|
0.464
|
NM_001692
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1
|
|
chr5_+_179247911
|
0.456
|
|
SQSTM1
|
sequestosome 1
|
|
chr3_+_52489617
|
0.456
|
|
NISCH
|
nischarin
|
|
chr5_+_179247903
|
0.452
|
|
SQSTM1
|
sequestosome 1
|
|
chr5_+_179247841
|
0.451
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr2_+_205410465
|
0.449
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr13_-_37633568
|
0.448
|
NM_001014286
NM_017569
|
FAM48A
|
family with sequence similarity 48, member A
|
|
chr16_-_57836370
|
0.447
|
|
KIFC3
|
kinesin family member C3
|
|
chr10_+_11865310
|
0.446
|
NM_153256
|
C10orf47
|
chromosome 10 open reading frame 47
|
|
chr22_+_19929310
|
0.445
|
|
COMT
|
catechol-O-methyltransferase
|
|
chr3_+_52280185
|
0.441
|
NM_001122870
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M
|
|
chr16_-_66959418
|
0.440
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr19_+_35532617
|
0.436
|
|
HPN
|
hepsin
|
|
chr16_-_57836380
|
0.435
|
NM_001130100
NM_005550
|
KIFC3
|
kinesin family member C3
|
|
chr22_-_51016343
|
0.435
|
NM_001145137
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle)
|
|
chr7_+_100081549
|
0.432
|
NM_173564
|
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1
|
|
chr11_+_61522860
|
0.429
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr22_+_19929262
|
0.429
|
NM_000754
|
COMT
|
catechol-O-methyltransferase
|
|
chr7_+_74508346
|
0.429
|
NM_001003795
|
GTF2IRD2B
GTF2IRD2
|
GTF2I repeat domain containing 2B
GTF2I repeat domain containing 2
|
|
chr5_+_179248089
|
0.427
|
|
SQSTM1
|
sequestosome 1
|
|
chr22_+_23412727
|
0.422
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr19_+_42829760
|
0.418
|
NM_001410
|
MEGF8
|
multiple EGF-like-domains 8
|
|
chr16_+_2034016
|
0.414
|
NM_005262
|
GFER
|
growth factor, augmenter of liver regeneration
|
|
chr7_-_148823388
|
0.412
|
NM_001001661
|
ZNF425
|
zinc finger protein 425
|
|
chr19_-_11373117
|
0.411
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chrX_+_53111482
|
0.405
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr17_+_79651072
|
0.404
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr3_-_52001388
|
0.403
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr16_-_75272524
|
0.403
|
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr19_-_36247917
|
0.401
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr5_-_81046858
|
0.401
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr5_+_1008900
|
0.396
|
NM_033120
|
NKD2
|
naked cuticle homolog 2 (Drosophila)
|
|
chr3_-_172166202
|
0.395
|
NM_004122
NM_198407
|
GHSR
|
growth hormone secretagogue receptor
|
|
chr2_+_149632836
|
0.394
|
|
KIF5C
|
kinesin family member 5C
|
|
chr6_+_29894105
|
0.394
|
|
|
|
|
chr9_+_130830447
|
0.394
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr19_+_35550794
|
0.394
|
|
HPN
|
hepsin
|
|
chr1_+_32827797
|
0.393
|
NM_052841
|
TSSK3
|
testis-specific serine kinase 3
|
|
chr15_-_60884615
|
0.393
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr2_-_85641128
|
0.392
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr13_+_42614171
|
0.387
|
NM_001204504
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr5_+_176513819
|
0.383
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr1_-_21058701
|
0.381
|
|
SH2D5
|
SH2 domain containing 5
|
|
chr17_-_48207113
|
0.379
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr5_-_108063960
|
0.379
|
|
HP07349
|
uncharacterized protein HP07349
|
|
chr19_-_12886263
|
0.379
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr7_+_6655526
|
0.378
|
NM_017560
|
ZNF853
|
zinc finger protein 853
|
|
chr22_+_39853316
|
0.375
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr6_-_154831704
|
0.375
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr6_-_31846766
|
0.373
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr7_+_73868111
|
0.373
|
NM_001199207
NM_005685
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr15_+_74908195
|
0.372
|
|
CLK3
|
CDC-like kinase 3
|
|
chr16_+_619905
|
0.371
|
NM_004204
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q
|
|
chr19_-_56110851
|
0.370
|
NM_032836
|
FIZ1
|
FLT3-interacting zinc finger 1
|
|
chr6_+_41514133
|
0.369
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr17_+_79651042
|
0.367
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr9_+_136287119
|
0.365
|
NM_139025
NM_139026
NM_139027
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr13_-_108519459
|
0.365
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr11_+_109964086
|
0.364
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr17_+_17206626
|
0.363
|
NM_020201
|
NT5M
|
5',3'-nucleotidase, mitochondrial
|
|
chr9_+_130830595
|
0.360
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr20_-_62711160
|
0.357
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr6_-_33266700
|
0.355
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr22_-_24096551
|
0.354
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr6_-_150039219
|
0.346
|
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr19_+_15218141
|
0.344
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr18_+_55711388
|
0.342
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr11_+_74459900
|
0.340
|
NM_001098638
|
RNF169
|
ring finger protein 169
|
|
chr9_+_35490018
|
0.340
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_128312345
|
0.339
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr7_-_2272575
|
0.338
|
NM_001013836
NM_001013837
NM_003550
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast)
|
|
chr1_-_11866079
|
0.338
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr7_-_2272552
|
0.338
|
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast)
|
|
chr3_+_77089248
|
0.338
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr1_+_16010816
|
0.337
|
NM_015164
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr22_-_50964573
|
0.337
|
NM_001169110
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr6_-_154831652
|
0.336
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr9_-_35111534
|
0.334
|
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr12_+_132413776
|
0.334
|
NM_001002019
NM_025215
|
PUS1
|
pseudouridylate synthase 1
|
|
chr19_+_55795533
|
0.333
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr7_-_1980141
|
0.328
|
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast)
|
|
chr1_+_36621457
|
0.328
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr7_-_33148897
|
0.327
|
NM_203288
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant)
|
|
chr16_+_87993476
|
0.327
|
NM_001173542
|
BANP
|
BTG3 associated nuclear protein
|
|
chr11_+_67034164
|
0.327
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr8_+_145582208
|
0.326
|
NM_001253816
NM_001253815
|
GPR172A
|
G protein-coupled receptor 172A
|
|
chr2_+_205410716
|
0.326
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr6_+_88117689
|
0.326
|
NM_001031743
|
C6orf165
|
chromosome 6 open reading frame 165
|
|
chr9_+_35489989
|
0.325
|
NM_014806
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr17_-_37307694
|
0.325
|
|
PLXDC1
|
plexin domain containing 1
|
|
chr5_-_132166589
|
0.324
|
NM_001172700
|
SHROOM1
|
shroom family member 1
|
|
chr3_-_50383009
|
0.321
|
NM_015896
|
ZMYND10
|
zinc finger, MYND-type containing 10
|
|
chr9_-_35748875
|
0.321
|
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr5_+_167956545
|
0.320
|
|
|
|
|
chr6_-_154831788
|
0.320
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr16_-_21531694
|
0.319
|
|
SLC7A5P2
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 pseudogene 2
|
|
chr8_-_145018904
|
0.319
|
NM_201381
|
PLEC
|
plectin
|
|
chr12_+_57482922
|
0.318
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr17_+_73008744
|
0.317
|
NM_001545
|
ICT1
|
immature colon carcinoma transcript 1
|
|
chr19_+_36705503
|
0.317
|
NM_001099638
NM_001099639
NM_007145
|
ZNF146
|
zinc finger protein 146
|
|
chr6_+_7541837
|
0.316
|
|
DSP
|
desmoplakin
|
|
chr9_-_35749144
|
0.316
|
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr20_+_48429375
|
0.316
|
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr10_+_102758072
|
0.316
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr6_-_150039391
|
0.315
|
NM_004690
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr11_+_67034148
|
0.315
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr6_+_7541863
|
0.313
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr2_-_20850822
|
0.310
|
NM_022460
|
HS1BP3
|
HCLS1 binding protein 3
|
|
chr19_-_4535202
|
0.310
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr14_+_105173953
|
0.310
|
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr19_+_47249302
|
0.309
|
NM_001039885
NM_024301
|
FKRP
|
fukutin related protein
|
|
chr6_+_7541892
|
0.309
|
|
DSP
|
desmoplakin
|
|
chr17_-_80291903
|
0.309
|
NM_003004
|
SECTM1
|
secreted and transmembrane 1
|
|
chr7_-_149321658
|
0.306
|
|
ZNF767
|
zinc finger family member 767
|
|
chr11_+_394199
|
0.304
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr19_-_46366337
|
0.304
|
NM_004819
|
SYMPK
|
symplekin
|
|
chr3_-_197463713
|
0.304
|
NM_014687
|
KIAA0226
|
KIAA0226
|
|
chr4_+_4861371
|
0.303
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr19_+_56154973
|
0.301
|
NM_016535
|
ZNF581
|
zinc finger protein 581
|
|
chr7_+_73868236
|
0.300
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr12_+_57482886
|
0.300
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr6_-_11232883
|
0.299
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr10_-_98945662
|
0.298
|
NM_003061
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr2_+_205410556
|
0.297
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr2_-_134179325
|
0.297
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr11_+_61447816
|
0.295
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr9_-_138987096
|
0.294
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr15_-_64126133
|
0.292
|
NM_003922
|
HERC1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1
|
|
chr12_-_112036257
|
0.292
|
|
ATXN2
|
ataxin 2
|
|
chr9_-_110251308
|
0.291
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_-_221915458
|
0.291
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr19_+_12305829
|
0.290
|
|
LOC100289333
|
uncharacterized LOC100289333
|
|
chr7_+_1581072
|
0.290
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr19_-_10230534
|
0.290
|
NM_003755
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G
|
|
chr12_-_101603984
|
0.290
|
NM_145913
|
SLC5A8
|
solute carrier family 5 (iodide transporter), member 8
|
|
chr9_-_131940481
|
0.289
|
|
IER5L
|
immediate early response 5-like
|
|
chr12_-_112036867
|
0.289
|
|
ATXN2
|
ataxin 2
|
|
chr13_+_78271820
|
0.288
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr5_-_54529424
|
0.288
|
NM_021147
|
CCNO
|
cyclin O
|
|
chr1_+_16083132
|
0.287
|
|
FBLIM1
|
filamin binding LIM protein 1
|