|
chr8_-_17658711
|
1.432
|
|
|
|
|
chr15_-_83621350
|
0.934
|
NM_004839
NM_199330
|
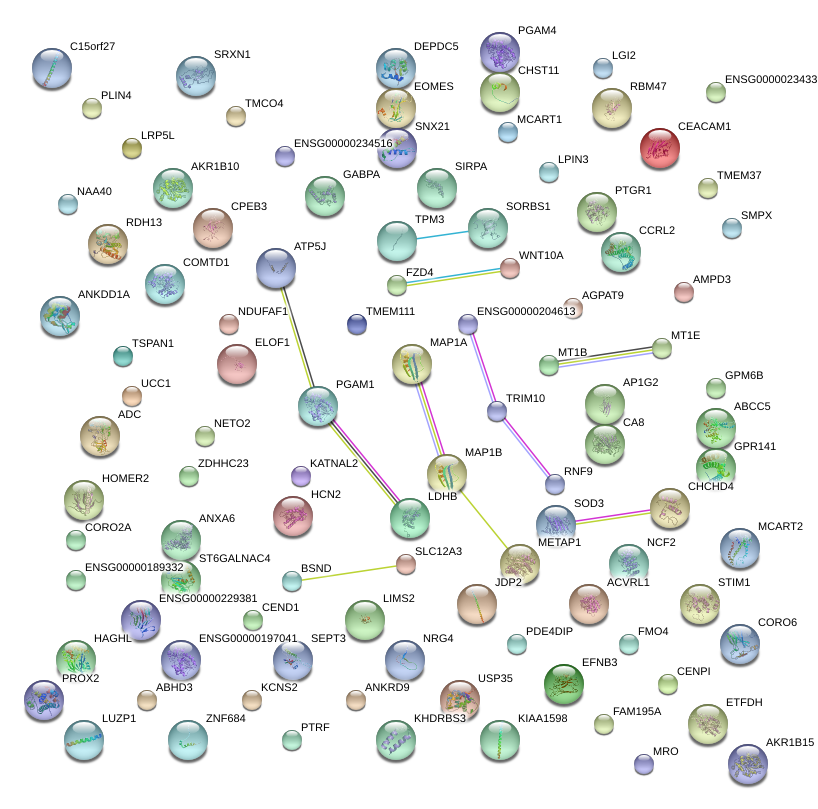
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr15_+_43809805
|
0.746
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr20_-_633808
|
0.592
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr20_-_633844
|
0.572
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chrX_-_21776158
|
0.544
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr15_+_76352270
|
0.433
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr1_-_183560010
|
0.424
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr9_-_114362100
|
0.423
|
NM_001146108
NM_001146109
|
PTGR1
|
prostaglandin reductase 1
|
|
chr4_+_84457390
|
0.416
|
NM_032717
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr7_+_134212343
|
0.413
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr1_+_171283372
|
0.377
|
NM_002022
|
FMO4
|
flavin containing monooxygenase 4
|
|
chr7_+_37960162
|
0.368
|
NM_001242946
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr20_+_39969420
|
0.365
|
NM_022896
|
LPIN3
|
lipin 3
|
|
chr14_-_102976096
|
0.361
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr9_-_130679210
|
0.360
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr12_-_21810720
|
0.358
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr14_+_88490893
|
0.354
|
|
LOC283587
|
uncharacterized LOC283587
|
|
chr22_+_42372763
|
0.351
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr1_-_144994839
|
0.348
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr4_+_24797084
|
0.344
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr11_-_790103
|
0.337
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr2_-_128432642
|
0.325
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr11_+_65190272
|
0.321
|
|
|
|
|
chr16_+_776957
|
0.321
|
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr4_+_159593233
|
0.317
|
NM_004453
|
ETFDH
|
electron-transferring-flavoprotein dehydrogenase
|
|
chr16_-_47177902
|
0.316
|
NM_001201477
NM_018092
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr16_-_47177832
|
0.314
|
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr11_-_86666211
|
0.313
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr11_+_65190267
|
0.310
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr12_-_21810754
|
0.303
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr14_-_102975996
|
0.302
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr1_-_154164551
|
0.296
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr20_+_44462788
|
0.289
|
|
SNX21
|
sorting nexin family member 21
|
|
chr15_-_76352068
|
0.286
|
|
NRG4
|
neuregulin 4
|
|
chr1_+_55464605
|
0.280
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr5_+_71403040
|
0.280
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr10_-_76995685
|
0.278
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr2_+_219745243
|
0.275
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chrX_+_100354797
|
0.272
|
NM_006733
|
CENPI
|
centromere protein I
|
|
chr4_-_25032306
|
0.272
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr4_+_24797203
|
0.268
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr6_-_30128688
|
0.268
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr1_-_20126368
|
0.265
|
NM_181719
|
TMCO4
|
transmembrane and coiled-coil domains 4
|
|
chr9_-_100954883
|
0.264
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr15_+_65204053
|
0.262
|
NM_182703
|
ANKDD1A
|
ankyrin repeat and death domain containing 1A
|
|
chr15_-_41694619
|
0.261
|
NM_016013
|
NDUFAF1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1
|
|
chr17_+_7608413
|
0.252
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr22_-_25758523
|
0.248
|
NM_182492
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like
|
|
chr11_+_77899857
|
0.245
|
NM_020798
|
USP35
|
ubiquitin specific peptidase 35
|
|
chr10_-_118765087
|
0.244
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr18_+_44497512
|
0.242
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr16_+_691901
|
0.242
|
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr19_-_4517715
|
0.239
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr3_-_183735617
|
0.239
|
NM_001023587
NM_005688
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr22_+_32149936
|
0.236
|
NM_001242897
|
DEPDC5
|
DEP domain containing 5
|
|
chr4_-_40517913
|
0.236
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr20_+_1875825
|
0.236
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr8_+_136469557
|
0.235
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr1_+_46640748
|
0.235
|
NM_005727
|
TSPAN1
|
tetraspanin 1
|
|
chr12_+_52306112
|
0.234
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr12_-_21810769
|
0.231
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr11_+_10472223
|
0.231
|
NM_000480
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr19_+_28284376
|
0.230
|
|
|
|
|
chr3_-_27763784
|
0.230
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr18_-_48351732
|
0.230
|
NM_001127174
|
MRO
|
maestro
|
|
chr18_-_19284737
|
0.230
|
NM_138340
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr5_-_139780654
|
0.228
|
|
|
|
|
chr12_+_105153258
|
0.225
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr18_-_19284622
|
0.225
|
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr2_+_120189342
|
0.224
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr3_-_14166228
|
0.217
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr1_-_23504273
|
0.215
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr1_+_33546700
|
0.214
|
NM_052998
|
ADC
|
arginine decarboxylase
|
|
chr3_-_14166370
|
0.212
|
NM_001098502
NM_144636
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr10_+_99186012
|
0.211
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr10_+_99186040
|
0.210
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr9_-_37904331
|
0.210
|
NM_033412
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr10_+_99186042
|
0.210
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr11_+_63706433
|
0.209
|
NM_024771
|
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae)
|
|
chr19_+_589849
|
0.208
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr10_+_99186017
|
0.207
|
NM_002629
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr8_-_61193882
|
0.207
|
NM_004056
|
CA8
|
carbonic anhydrase VIII
|
|
chr3_-_14166246
|
0.206
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr22_+_32149962
|
0.204
|
|
DEPDC5
|
DEP domain containing 5
|
|
chr12_-_21810733
|
0.203
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810779
|
0.201
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr3_+_113666544
|
0.196
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr10_+_99186004
|
0.194
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr11_+_3876919
|
0.193
|
NM_003156
|
STIM1
|
stromal interaction molecule 1
|
|
chr11_+_10524398
|
0.191
|
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr14_-_75330535
|
0.191
|
NM_001080408
NM_001243007
|
PROX2
|
prospero homeobox 2
|
|
chr17_-_40575117
|
0.191
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr17_-_27948345
|
0.191
|
NM_032854
|
CORO6
|
coronin 6
|
|
chr1_+_40997232
|
0.190
|
NM_152373
|
ZNF684
|
zinc finger protein 684
|
|
chrX_-_13835187
|
0.186
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_40997290
|
0.185
|
|
ZNF684
|
zinc finger protein 684
|
|
chr5_-_150537261
|
0.184
|
|
ANXA6
|
annexin A6
|
|
chr14_+_75898836
|
0.183
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr1_-_144994908
|
0.181
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr21_+_27107257
|
0.181
|
NM_001197297
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr3_-_10028344
|
0.181
|
|
TMEM111
|
transmembrane protein 111
|
|
chr21_-_27107184
|
0.181
|
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6
|
|
chr19_-_43031343
|
0.178
|
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr20_+_54933982
|
0.178
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr3_+_46448678
|
0.178
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr19_-_55574434
|
0.174
|
NM_001145971
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr16_+_56899118
|
0.174
|
NM_000339
NM_001126107
NM_001126108
|
SLC12A3
|
solute carrier family 12 (sodium/chloride transporters), member 3
|
|
chr8_+_99439249
|
0.174
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr3_-_14166292
|
0.173
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr5_-_150537306
|
0.169
|
|
ANXA6
|
annexin A6
|
|
chr10_-_94050799
|
0.169
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr14_-_24036532
|
0.167
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr16_+_56685810
|
0.165
|
NM_005947
|
MT1B
|
metallothionein 1B
|
|
chr22_+_32150847
|
0.164
|
NM_001136029
|
DEPDC5
|
DEP domain containing 5
|
|
chr3_-_197354629
|
0.164
|
|
LOC220729
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) pseudogene
|
|
chr5_-_150537335
|
0.164
|
NM_001155
|
ANXA6
|
annexin A6
|
|
chr12_-_46121489
|
0.164
|
|
LOC400027
|
uncharacterized LOC400027
|
|
chr10_-_97200895
|
0.163
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr1_+_29213619
|
0.163
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_+_16330730
|
0.162
|
NM_178840
|
C1orf64
|
chromosome 1 open reading frame 64
|
|
chr16_+_691827
|
0.160
|
NM_138418
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr9_-_15307325
|
0.160
|
NM_001168339
NM_001168340
NM_001168341
NM_152574
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr3_-_10028512
|
0.160
|
NM_018447
|
TMEM111
|
transmembrane protein 111
|
|
chr22_+_24199058
|
0.159
|
NM_001024938
NM_030807
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11
|
|
chr19_-_1512997
|
0.159
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr9_+_130965668
|
0.158
|
|
DNM1
|
dynamin 1
|
|
chr1_+_65210724
|
0.158
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr9_-_136344182
|
0.157
|
NM_001145099
NM_017585
|
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chrX_+_23801431
|
0.156
|
|
SAT1
|
spermidine/spermine N1-acetyltransferase 1
|
|
chr5_+_176784947
|
0.154
|
|
|
|
|
chr3_+_195384918
|
0.153
|
|
SDHAP1
SDHAP2
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 1
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2
|
|
chr6_+_136172802
|
0.153
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr12_-_772525
|
0.151
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr11_+_63993720
|
0.150
|
NM_001128612
NM_001128613
NM_032344
|
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22
|
|
chr12_+_25348194
|
0.150
|
|
LYRM5
|
LYR motif containing 5
|
|
chr4_+_56262120
|
0.150
|
|
TMEM165
|
transmembrane protein 165
|
|
chr17_-_48264039
|
0.149
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr9_+_95858449
|
0.149
|
NM_032310
|
C9orf89
|
chromosome 9 open reading frame 89
|
|
chr1_-_155178968
|
0.148
|
|
THBS3
|
thrombospondin 3
|
|
chr1_+_46769336
|
0.148
|
NM_006004
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein
|
|
chr2_-_176046120
|
0.147
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr10_+_105344714
|
0.147
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr14_+_105714853
|
0.147
|
NM_033271
|
BTBD6
|
BTB (POZ) domain containing 6
|
|
chr21_+_27107322
|
0.146
|
NM_002040
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr14_-_31889736
|
0.146
|
NM_015473
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chr9_+_103189494
|
0.146
|
NM_001198807
NM_080655
NM_001198805
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr3_+_13521714
|
0.145
|
NM_024827
NM_001136041
|
HDAC11
|
histone deacetylase 11
|
|
chr22_+_20105095
|
0.144
|
|
RANBP1
|
RAN binding protein 1
|
|
chr22_+_24891114
|
0.143
|
NM_016327
|
UPB1
|
ureidopropionase, beta
|
|
chr2_-_87088993
|
0.143
|
NM_001178100
NM_004931
NM_172101
NM_172102
NM_172213
|
CD8B
|
CD8b molecule
|
|
chr8_-_127570465
|
0.142
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr6_+_3259134
|
0.141
|
NM_001128591
NM_001128592
NM_001135750
|
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4
|
|
chr10_-_5060191
|
0.141
|
NM_001354
NM_205845
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr15_+_99191671
|
0.141
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr16_-_11680146
|
0.140
|
NM_004862
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr5_-_137071703
|
0.139
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr1_-_35325335
|
0.139
|
NM_001164825
NM_138428
|
C1orf212
|
chromosome 1 open reading frame 212
|
|
chr11_+_99426872
|
0.139
|
NM_175566
|
CNTN5
|
contactin 5
|
|
chr1_-_173886401
|
0.138
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr1_-_200992824
|
0.137
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr10_+_695887
|
0.136
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr10_+_99185977
|
0.135
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr18_-_44497307
|
0.135
|
NM_004671
NM_173206
|
PIAS2
|
protein inhibitor of activated STAT, 2
|
|
chr7_+_139208673
|
0.133
|
NM_001080511
|
CLEC2L
|
C-type lectin domain family 2, member L
|
|
chr15_-_34659245
|
0.133
|
NM_153613
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4
|
|
chr19_+_57742376
|
0.132
|
NM_001015879
NM_003160
NM_001015878
|
AURKC
|
aurora kinase C
|
|
chr20_+_1876006
|
0.132
|
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr15_-_23378167
|
0.131
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr2_-_42721083
|
0.131
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr1_-_173886464
|
0.131
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr8_+_124428964
|
0.130
|
NM_018024
|
WDYHV1
|
WDYHV motif containing 1
|
|
chr16_-_67427335
|
0.130
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr11_-_86383355
|
0.130
|
NM_006680
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chrX_-_129299680
|
0.130
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr3_+_73046081
|
0.129
|
NM_174907
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr1_-_230849863
|
0.129
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr9_-_97356007
|
0.128
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chr2_+_242127923
|
0.128
|
NM_001001666
NM_001001891
|
ANO7
|
anoctamin 7
|
|
chr1_-_230849847
|
0.128
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr22_+_20105076
|
0.128
|
|
RANBP1
|
RAN binding protein 1
|
|
chr2_+_182850550
|
0.128
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr14_+_57735605
|
0.128
|
NM_018229
|
MUDENG
|
MU-2/AP1M2 domain containing, death-inducing
|
|
chr5_-_126409171
|
0.128
|
NM_001164478
NM_001164479
|
C5orf63
|
chromosome 5 open reading frame 63
|
|
chr5_-_72744351
|
0.127
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chrX_-_46618471
|
0.126
|
NM_032591
|
SLC9A7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7
|
|
chr9_+_130965666
|
0.126
|
|
DNM1
|
dynamin 1
|
|
chr6_-_46293628
|
0.124
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr4_-_40632639
|
0.124
|
|
RBM47
|
RNA binding motif protein 47
|
|
chrX_-_129299803
|
0.124
|
NM_001130847
NM_004208
NM_145812
NM_145813
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr1_+_29241002
|
0.123
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr9_+_130965608
|
0.123
|
NM_001005336
NM_004408
|
DNM1
|
dynamin 1
|
|
chr2_+_228678556
|
0.122
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr6_+_125475334
|
0.122
|
NM_001003395
|
TPD52L1
|
tumor protein D52-like 1
|
|
chr1_-_144994943
|
0.121
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr12_-_56753856
|
0.121
|
|
STAT2
|
signal transducer and activator of transcription 2, 113kDa
|
|
chr3_+_181417387
|
0.121
|
|
SOX2-OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr4_-_170947428
|
0.121
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr3_+_73045977
|
0.120
|
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr22_+_20105145
|
0.120
|
|
RANBP1
|
RAN binding protein 1
|