|
chr17_-_70588942
|
0.268
|
|
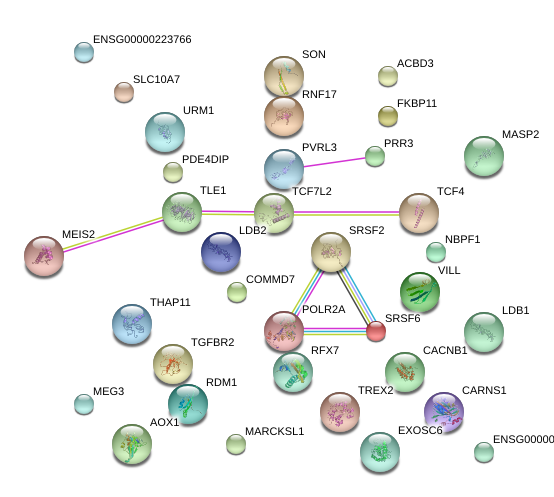
LOC100499467
|
uncharacterized LOC100499467
|
|
chr1_-_144994908
|
0.078
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_32801606
|
0.059
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr9_+_131133597
|
0.051
|
NM_001135947
NM_030914
|
URM1
|
ubiquitin related modifier 1
|
|
chr9_+_131133637
|
0.051
|
|
URM1
|
ubiquitin related modifier 1
|
|
chr1_-_144994839
|
0.029
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_11107295
|
0.027
|
NM_006610
NM_139208
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr14_+_101292451
|
0.026
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr2_+_201450579
|
0.021
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr18_-_53303129
|
0.021
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr16_-_70285794
|
0.020
|
NM_058219
|
EXOSC6
|
exosome component 6
|
|
chr1_-_11107150
|
0.017
|
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr2_+_201450730
|
0.016
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr4_-_147442868
|
0.013
|
NM_001029998
NM_032128
|
SLC10A7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7
|
|
chr17_-_34257730
|
0.011
|
NM_001034836
NM_001163120
NM_001163121
NM_145654
|
RDM1
|
RAD52 motif 1
|
|
chr20_-_31331196
|
0.011
|
|
COMMD7
|
COMM domain containing 7
|
|
chr1_-_144994943
|
0.011
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr3_+_38032260
|
0.008
|
|
VILL
|
villin-like
|
|
chr6_+_30524662
|
0.008
|
|
PRR3
|
proline rich 3
|
|
chr11_+_67183144
|
0.007
|
NM_001166222
NM_020811
|
CARNS1
|
carnosine synthase 1
|
|
chr17_-_74733241
|
0.007
|
|
SRSF2
|
serine/arginine-rich splicing factor 2
|
|
chr10_-_103879933
|
0.007
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr20_+_42086533
|
0.006
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr9_-_84304378
|
0.006
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr15_-_37393364
|
0.006
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr15_-_56535475
|
0.005
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr16_+_67876381
|
0.004
|
|
THAP11
|
THAP domain containing 11
|
|
chr17_+_7387688
|
0.003
|
NM_000937
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa
|
|
chr3_+_30648372
|
0.002
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr3_+_30647901
|
0.002
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr1_-_226374400
|
0.001
|
|
ACBD3
|
acyl-CoA binding domain containing 3
|
|
chr1_-_226374401
|
0.000
|
NM_022735
|
ACBD3
|
acyl-CoA binding domain containing 3
|
|
chr21_+_34922923
|
0.000
|
|
SON
|
SON DNA binding protein
|