|
chr16_+_72088507
|
3.261
|
NM_001126102
NM_005143
|
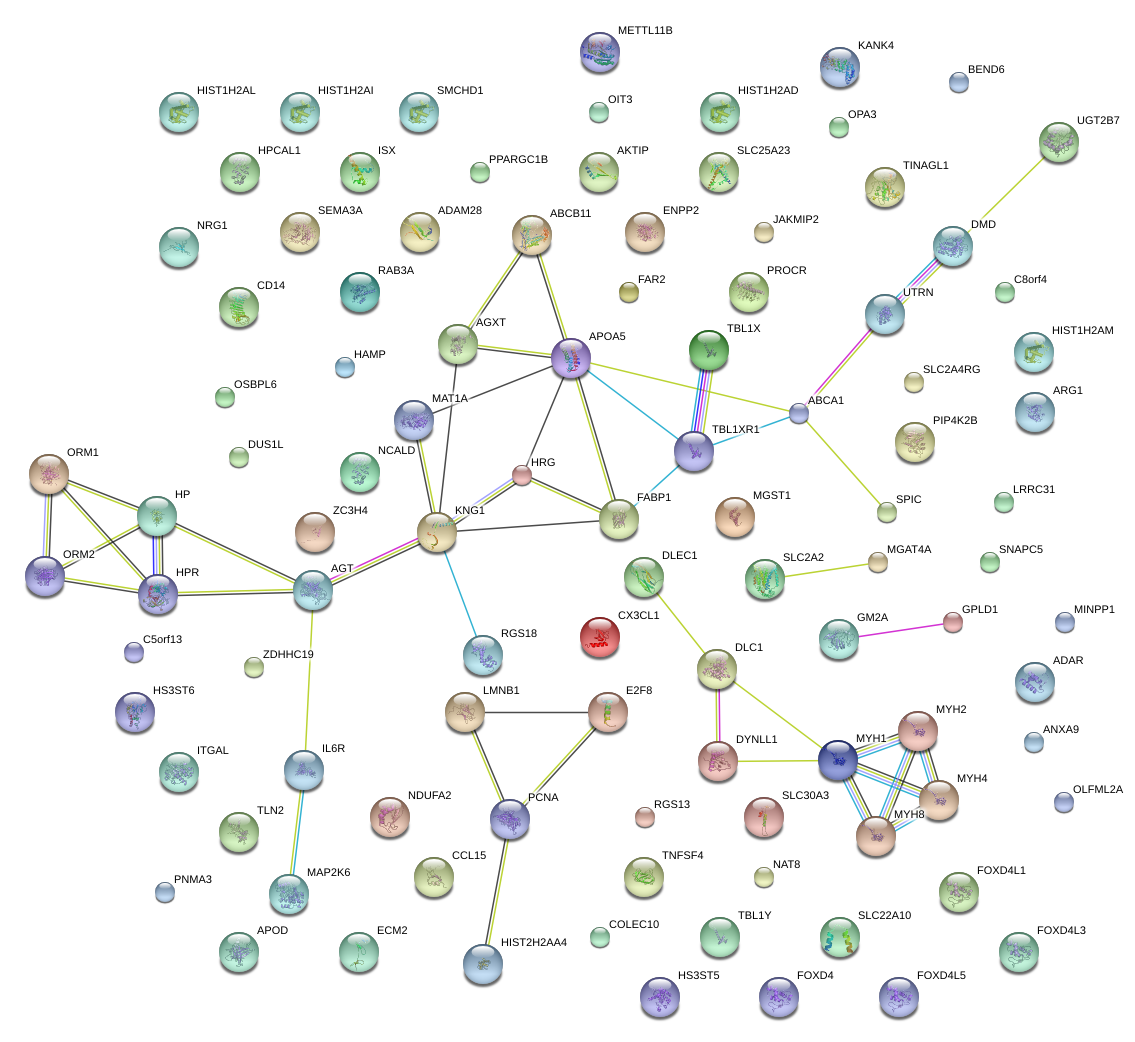
HP
|
haptoglobin
|
|
chr16_+_72097124
|
3.171
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr19_+_35773409
|
2.551
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr3_+_186383770
|
2.417
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr9_-_107666046
|
1.670
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_-_230849936
|
1.602
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr16_+_30484049
|
1.593
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr5_+_126112314
|
1.466
|
NM_005573
|
LMNB1
|
lamin B1
|
|
chr2_-_99279935
|
1.454
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr16_+_30483959
|
1.382
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr12_-_46121248
|
1.373
|
|
LOC400027
|
uncharacterized LOC400027
|
|
chr8_+_24151579
|
1.300
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr6_+_131894343
|
1.280
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr1_-_230849863
|
1.232
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr3_+_186435097
|
1.217
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr17_-_34328530
|
1.103
|
|
CCL14-CCL15
CCL15
|
CCL14-CCL15 readthrough
chemokine (C-C motif) ligand 15
|
|
chr9_+_117085302
|
1.081
|
NM_000607
|
ORM1
|
orosomucoid 1
|
|
chr6_-_24489738
|
1.077
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chr9_+_117092068
|
1.058
|
NM_000608
|
ORM2
|
orosomucoid 2
|
|
chr5_-_147162251
|
1.056
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr11_+_63057371
|
1.055
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chrX_+_152224862
|
1.033
|
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr8_-_120651019
|
1.012
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr1_-_230849847
|
1.009
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr3_-_170744458
|
0.980
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr6_+_27833033
|
0.952
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr1_+_150954498
|
0.936
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr10_-_82049178
|
0.927
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr1_-_173174611
|
0.854
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr2_-_73869493
|
0.831
|
NM_003960
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative)
|
|
chr5_-_140012730
|
0.825
|
|
CD14
|
CD14 molecule
|
|
chr12_-_46121489
|
0.818
|
|
LOC400027
|
uncharacterized LOC400027
|
|
chr3_-_190580464
|
0.809
|
NM_001146686
|
GMNC
|
geminin coiled-coil domain containing
|
|
chr11_-_116663106
|
0.800
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr19_-_18314731
|
0.795
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr8_+_40010986
|
0.792
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr1_-_62785067
|
0.790
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chrX_+_152224765
|
0.785
|
NM_013364
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr6_+_56819923
|
0.773
|
|
BEND6
|
BEN domain containing 6
|
|
chr8_+_120079423
|
0.764
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr5_+_150632612
|
0.742
|
NM_000405
NM_001167607
|
GM2A
|
GM2 ganglioside activator
|
|
chr17_-_36956145
|
0.740
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr17_-_10372875
|
0.713
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr3_-_195938256
|
0.697
|
NM_001039617
|
ZDHHC19
|
zinc finger, DHHC-type containing 19
|
|
chr9_-_95298251
|
0.694
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr2_+_179184970
|
0.689
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr15_+_63050761
|
0.676
|
|
TLN2
|
talin 2
|
|
chr6_-_114384008
|
0.675
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr3_-_169587598
|
0.664
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr19_-_47617008
|
0.657
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr6_+_34216344
|
0.653
|
|
|
|
|
chr20_-_5100443
|
0.644
|
NM_182649
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr17_+_67498391
|
0.642
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr1_+_154377847
|
0.635
|
|
IL6R
|
interleukin 6 receptor
|
|
chr22_+_35462125
|
0.629
|
NM_001008494
|
ISX
|
intestine-specific homeobox
|
|
chr2_-_169887832
|
0.599
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr5_+_149151502
|
0.598
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr19_-_46088074
|
0.588
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr3_-_12586962
|
0.587
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr8_-_103137134
|
0.566
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chrX_-_33229428
|
0.558
|
NM_004006
|
DMD
|
dystrophin
|
|
chr12_+_29376596
|
0.552
|
NM_018099
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr9_-_118416
|
0.550
|
NM_207305
|
FOXD4
|
forkhead box D4
|
|
chr2_-_88427535
|
0.545
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chrX_+_9431314
|
0.527
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr19_-_6459735
|
0.524
|
NM_024103
|
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23
|
|
chr20_+_62372436
|
0.523
|
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr16_+_57406398
|
0.518
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr10_+_74653338
|
0.517
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr3_-_195306274
|
0.515
|
|
APOD
|
apolipoprotein D
|
|
chr15_-_66790068
|
0.514
|
NM_006049
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa
|
|
chr5_-_147162264
|
0.512
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr5_-_111091947
|
0.499
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_+_89267589
|
0.495
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr1_-_62784971
|
0.493
|
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr12_+_16500075
|
0.491
|
NM_145792
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr7_-_83824216
|
0.484
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr20_+_33759773
|
0.484
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr4_+_69962191
|
0.483
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr1_-_154600446
|
0.476
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr1_+_192605267
|
0.471
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr17_-_34328607
|
0.463
|
|
CCL14-CCL15
|
CCL14-CCL15 readthrough
|
|
chr16_-_53537112
|
0.457
|
NM_001012398
NM_022476
|
AKTIP
|
AKT interacting protein
|
|
chr17_-_34329083
|
0.457
|
NM_032965
|
CCL15
|
chemokine (C-C motif) ligand 15
|
|
chr17_-_41277409
|
0.453
|
NM_007297
NM_007299
NM_007294
NM_007300
|
BRCA1
|
breast cancer 1, early onset
|
|
chr16_+_19079220
|
0.450
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr21_+_45079375
|
0.446
|
NM_015056
|
RRP1B
|
ribosomal RNA processing 1 homolog B (S. cerevisiae)
|
|
chr5_-_58882274
|
0.446
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr16_+_31711914
|
0.446
|
|
KIAA0664L3
|
KIAA0664-like 3
|
|
chr5_+_36606456
|
0.441
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr19_-_35992798
|
0.440
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr5_-_53606387
|
0.436
|
NM_019087
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr20_+_42187634
|
0.436
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr4_+_108911008
|
0.430
|
|
HADH
|
hydroxyacyl-CoA dehydrogenase
|
|
chr1_-_154600383
|
0.427
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr5_+_36606667
|
0.423
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr7_-_15601574
|
0.423
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr12_-_8043743
|
0.418
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr7_-_45026204
|
0.410
|
|
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding)
|
|
chr14_+_75894508
|
0.405
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr1_+_229440128
|
0.394
|
NM_006542
|
SPHAR
|
S-phase response (cyclin related)
|
|
chr15_-_42840910
|
0.393
|
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr16_+_68057144
|
0.388
|
NM_017803
|
DUS2L
|
dihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae)
|
|
chr2_-_110962539
|
0.386
|
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr2_-_191872322
|
0.384
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr15_-_23034365
|
0.383
|
|
NIPA2
|
non imprinted in Prader-Willi/Angelman syndrome 2
|
|
chr9_+_129724503
|
0.382
|
NM_001190728
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr3_+_101280679
|
0.382
|
NM_017819
|
RG9MTD1
|
RNA (guanine-9-) methyltransferase domain containing 1
|
|
chr1_+_151032150
|
0.380
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr10_+_91152321
|
0.379
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr5_+_42548314
|
0.374
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr17_-_41277304
|
0.370
|
|
BRCA1
|
breast cancer 1, early onset
|
|
chr14_-_23285011
|
0.360
|
NM_001126105
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr2_-_190627923
|
0.359
|
NM_022353
|
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1
|
|
chr1_+_40839368
|
0.359
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr3_-_185655833
|
0.350
|
NM_001243879
NM_004593
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr16_-_30457197
|
0.350
|
NM_012248
|
SEPHS2
|
selenophosphate synthetase 2
|
|
chr20_+_23342786
|
0.348
|
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr19_-_41356193
|
0.347
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr7_+_151722774
|
0.338
|
NM_022087
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chrX_-_117251302
|
0.337
|
NM_001168301
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr6_-_30080866
|
0.335
|
NM_007028
|
TRIM31
|
tripartite motif containing 31
|
|
chr17_-_41322211
|
0.331
|
|
BRCA1
|
breast cancer 1, early onset
|
|
chr11_-_6462129
|
0.329
|
NM_000613
|
HPX
|
hemopexin
|
|
chr13_-_41768533
|
0.328
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr6_-_30028755
|
0.328
|
|
ZNRD1-AS1
|
ZNRD1 antisense RNA 1 (non-protein coding)
|
|
chr7_+_94297448
|
0.326
|
|
PEG10
|
paternally expressed 10
|
|
chr14_-_23284617
|
0.326
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr9_-_77503009
|
0.326
|
NM_017662
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr19_-_49622278
|
0.325
|
NM_018111
|
C19orf73
|
chromosome 19 open reading frame 73
|
|
chr9_-_6015606
|
0.320
|
NM_001243202
NM_001243203
NM_012416
|
RANBP6
|
RAN binding protein 6
|
|
chr1_-_230850335
|
0.319
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr1_+_100111430
|
0.318
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr12_+_124773709
|
0.316
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr1_+_196857143
|
0.313
|
NM_001201550
NM_001201551
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr6_-_2840683
|
0.312
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr5_+_140718353
|
0.312
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr21_+_45138973
|
0.309
|
NM_003681
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chr2_+_219745243
|
0.308
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr9_+_111624545
|
0.308
|
NM_006687
|
ACTL7A
|
actin-like 7A
|
|
chr2_+_201756639
|
0.306
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae)
|
|
chr11_-_18062304
|
0.305
|
NM_004179
|
TPH1
|
tryptophan hydroxylase 1
|
|
chr3_-_185655806
|
0.304
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr5_+_95997871
|
0.303
|
NM_001042440
|
CAST
|
calpastatin
|
|
chr9_-_77643307
|
0.301
|
NM_152420
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr12_+_7013896
|
0.299
|
NM_001135217
NM_006992
NM_201650
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr19_+_36486042
|
0.294
|
NM_001042631
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1
|
|
chr5_+_42425093
|
0.294
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr10_+_99496938
|
0.292
|
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr3_+_122044010
|
0.292
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr1_-_109935869
|
0.291
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr6_-_56407681
|
0.290
|
|
|
|
|
chr8_+_38585703
|
0.287
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr17_+_74536114
|
0.286
|
NM_001077620
|
PRCD
|
progressive rod-cone degeneration
|
|
chr19_-_36233311
|
0.283
|
NM_024660
|
IGFLR1
|
IGF-like family receptor 1
|
|
chr20_+_23342844
|
0.278
|
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr2_-_152684882
|
0.277
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr1_-_173176314
|
0.277
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr15_-_63674074
|
0.276
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr3_+_119501556
|
0.274
|
NM_022002
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr19_-_46088021
|
0.273
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr3_-_185655776
|
0.270
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr3_+_155860750
|
0.268
|
NM_003471
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr1_+_40839429
|
0.266
|
|
SMAP2
|
small ArfGAP2
|
|
chr3_+_124303505
|
0.265
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr5_+_140729723
|
0.261
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr3_-_170744529
|
0.258
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr6_+_26204779
|
0.256
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr11_-_33795892
|
0.255
|
NM_012175
NM_033406
|
FBXO3
|
F-box protein 3
|
|
chr4_+_155484131
|
0.255
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr4_-_159092509
|
0.252
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_150399980
|
0.251
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr16_+_103009
|
0.250
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr1_+_119683006
|
0.245
|
|
|
|
|
chr6_+_131148544
|
0.245
|
NM_001195597
|
LOC100507203
|
uncharacterized LOC100507203
|
|
chr4_+_108911042
|
0.243
|
|
HADH
|
hydroxyacyl-CoA dehydrogenase
|
|
chr2_-_86422892
|
0.243
|
NM_001100169
NM_001100170
NM_006839
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr1_-_102462371
|
0.242
|
|
OLFM3
|
olfactomedin 3
|
|
chr7_-_129592711
|
0.239
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr3_-_185655648
|
0.239
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr6_+_150690027
|
0.238
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr8_+_98788014
|
0.238
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr16_-_30457168
|
0.236
|
|
SEPHS2
|
selenophosphate synthetase 2
|
|
chr15_+_91411884
|
0.234
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr4_-_104119475
|
0.233
|
NM_001813
|
CENPE
|
centromere protein E, 312kDa
|
|
chr11_-_68039327
|
0.232
|
|
C11orf24
|
chromosome 11 open reading frame 24
|
|
chr7_-_45808541
|
0.231
|
|
SEPT7P2
|
septin 7 pseudogene 2
|
|
chr9_-_77643179
|
0.230
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr20_+_42187705
|
0.230
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr2_-_158300517
|
0.230
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chrX_-_13835213
|
0.229
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_67519698
|
0.229
|
|
SLC35D1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1
|
|
chr6_-_116447225
|
0.229
|
NM_000493
|
COL10A1
|
collagen, type X, alpha 1
|
|
chr9_-_14398981
|
0.229
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_70083055
|
0.227
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr10_-_104211244
|
0.227
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr3_+_186330844
|
0.226
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr11_-_64647136
|
0.225
|
|
EHD1
|
EH-domain containing 1
|
|
chr7_+_56019552
|
0.224
|
NM_015969
|
MRPS17
|
mitochondrial ribosomal protein S17
|
|
chr12_+_65224214
|
0.223
|
|
|
|