|
chr19_+_45971249
|
11.992
|
NM_001114171
NM_006732
|
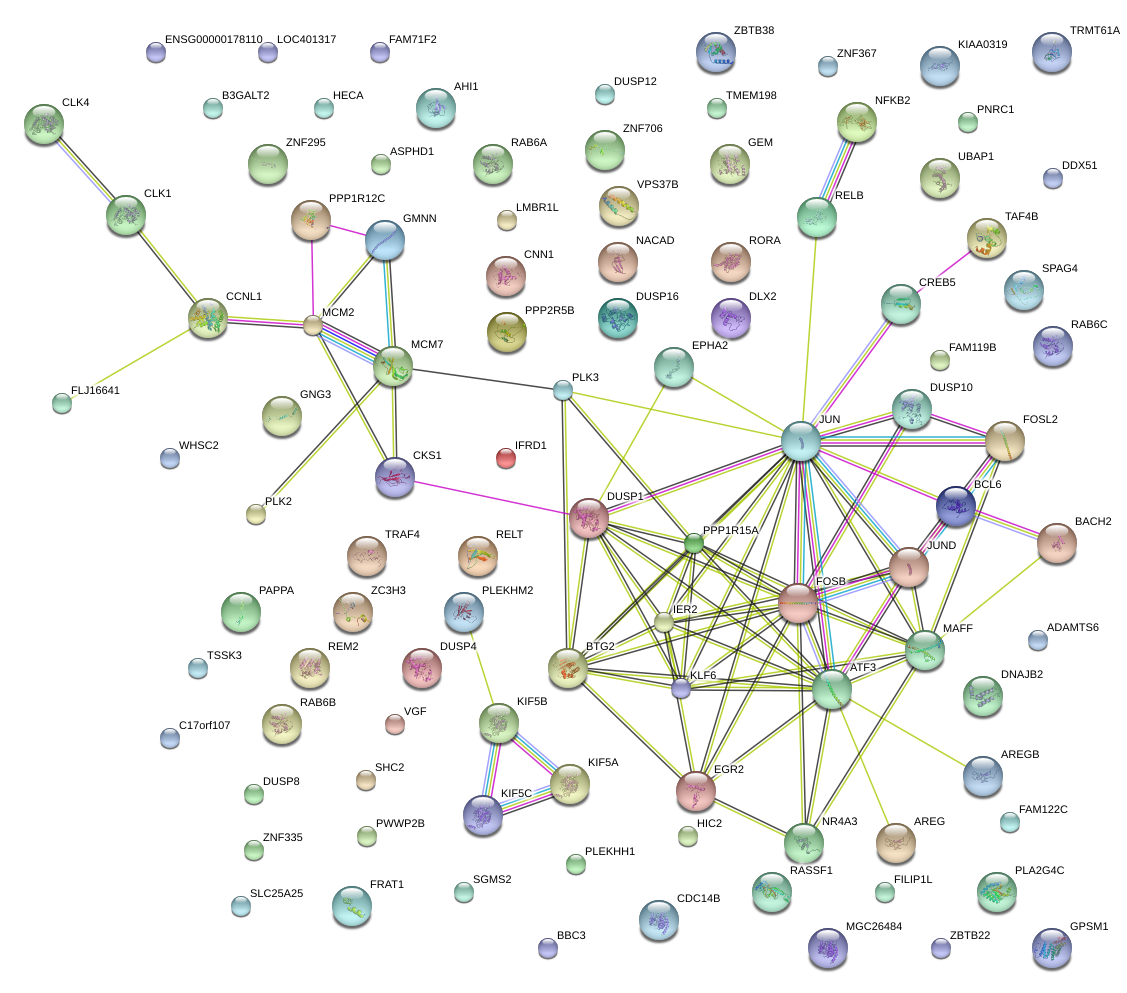
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr11_-_1593149
|
8.589
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr5_-_172198160
|
6.612
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr19_+_49375677
|
6.298
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr19_+_49375638
|
6.142
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr19_+_49375679
|
6.120
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr7_+_28448970
|
5.745
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr8_-_95274540
|
4.962
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr6_-_91006580
|
4.874
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr1_+_212782103
|
4.722
|
|
ATF3
|
activating transcription factor 3
|
|
chr12_-_123380682
|
4.627
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr22_+_38597938
|
4.560
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr6_-_91006460
|
4.526
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr12_-_123380608
|
4.513
|
|
|
|
|
chr4_+_75480628
|
4.412
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr8_-_95274520
|
4.092
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr12_-_123380628
|
3.948
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr6_+_139456240
|
3.892
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr19_+_45504686
|
3.890
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr22_+_38598065
|
3.804
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr8_+_128806720
|
3.659
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr1_-_59249687
|
3.522
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr19_+_11649578
|
3.426
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr6_+_135818938
|
3.402
|
|
LINC00271
|
long intergenic non-protein coding RNA 271
|
|
chr2_+_28615695
|
3.382
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_28615668
|
3.337
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_28615771
|
3.291
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_+_128312345
|
3.287
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr7_-_100808832
|
3.181
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr3_+_156544075
|
3.089
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr1_+_212738675
|
3.053
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr1_-_713985
|
2.900
|
|
LOC100288069
|
general transcription factor IIi pseudogene
|
|
chr21_-_43430448
|
2.775
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr2_+_220408742
|
2.435
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr14_+_23352430
|
2.262
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr1_-_221915393
|
2.261
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr7_+_28725597
|
2.225
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_-_3827418
|
2.188
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr1_-_221915426
|
2.183
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_-_221915458
|
2.080
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_+_32827797
|
2.074
|
NM_052841
|
TSSK3
|
testis-specific serine kinase 3
|
|
chr1_+_212781969
|
2.063
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr3_-_8543327
|
2.055
|
|
LOC100288428
|
uncharacterized LOC100288428
|
|
chr20_+_34203746
|
2.054
|
NM_003116
|
SPAG4
|
sperm associated antigen 4
|
|
chr12_-_12715244
|
2.033
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr3_-_50374875
|
2.029
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr10_-_3827205
|
2.027
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr5_-_57755787
|
2.018
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr9_+_130860769
|
2.015
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chrX_+_133941204
|
2.004
|
NM_001170782
NM_001170784
NM_001170779
NM_001170781
NM_001170783
NM_138819
|
FAM122C
|
family with sequence similarity 122C
|
|
chr2_+_149632772
|
2.001
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr18_+_23806385
|
1.916
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr19_-_460995
|
1.900
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr3_+_141087298
|
1.881
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr2_+_149632836
|
1.879
|
|
KIF5C
|
kinesin family member 5C
|
|
chr3_+_50297644
|
1.877
|
|
|
|
|
chr10_+_134210691
|
1.874
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr1_+_203274646
|
1.849
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr19_-_47734215
|
1.843
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr11_+_73087396
|
1.841
|
NM_152222
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr20_-_44600939
|
1.820
|
|
ZNF335
|
zinc finger protein 335
|
|
chr9_+_34179047
|
1.814
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr10_+_104154228
|
1.775
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr22_+_21771661
|
1.758
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr9_+_102584136
|
1.748
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr19_+_13263890
|
1.738
|
|
IER2
|
immediate early response 2
|
|
chr1_-_16482426
|
1.730
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr12_-_132628841
|
1.701
|
NM_175066
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr2_-_172967627
|
1.686
|
|
DLX2
|
distal-less homeobox 2
|
|
chr1_+_16010816
|
1.682
|
NM_015164
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr8_-_144623611
|
1.674
|
NM_015117
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr8_-_144623579
|
1.673
|
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr14_+_103995535
|
1.660
|
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr3_-_50374795
|
1.635
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr5_-_178054072
|
1.635
|
|
CLK4
|
CDC-like kinase 4
|
|
chr5_-_178054089
|
1.601
|
|
CLK4
|
CDC-like kinase 4
|
|
chr19_-_55628780
|
1.580
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C
|
|
chr4_+_108745718
|
1.567
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr17_+_27071008
|
1.564
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr2_+_220144024
|
1.549
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr3_-_99594915
|
1.535
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_-_193155723
|
1.528
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr3_-_156877882
|
1.522
|
|
CCNL1
|
cyclin L1
|
|
chr1_+_16010838
|
1.516
|
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr8_-_29207795
|
1.512
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr14_+_103995473
|
1.511
|
NM_152307
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr12_-_49504612
|
1.509
|
|
LMBR1L
|
limb region 1 homolog (mouse)-like
|
|
chr9_+_130860816
|
1.509
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr11_+_64692052
|
1.506
|
NM_006244
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta
|
|
chr10_-_64576074
|
1.505
|
|
EGR2
|
early growth response 2
|
|
chr6_+_89791510
|
1.483
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr10_-_64576123
|
1.483
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr15_-_60884615
|
1.464
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_-_99382073
|
1.461
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr1_+_45266035
|
1.448
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr6_-_135818786
|
1.444
|
|
AHI1
|
Abelson helper integration site 1
|
|
chr3_-_187463474
|
1.433
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_-_18392048
|
1.432
|
|
JUND
|
jun D proto-oncogene
|
|
chr14_+_67999915
|
1.428
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr5_-_64777703
|
1.425
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr19_-_48614013
|
1.414
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr10_+_99079017
|
1.408
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr7_-_45128492
|
1.399
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr16_+_29912146
|
1.398
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr20_-_44600787
|
1.389
|
NM_022095
|
ZNF335
|
zinc finger protein 335
|
|
chr17_+_4802866
|
1.384
|
NM_001145536
|
C17orf107
|
chromosome 17 open reading frame 107
|
|
chr11_+_62475113
|
1.378
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr4_-_2010961
|
1.374
|
NM_005663
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr7_+_112090463
|
1.372
|
NM_001550
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr9_+_139221931
|
1.359
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr3_-_133614296
|
1.359
|
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr19_-_42759279
|
1.357
|
NM_006494
|
ERF
|
Ets2 repressor factor
|
|
chr9_-_34397786
|
1.347
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr19_-_51014344
|
1.340
|
NM_138334
|
JOSD2
|
Josephin domain containing 2
|
|
chr15_+_78556901
|
1.330
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr19_-_59066372
|
1.319
|
|
CHMP2A
|
charged multivesicular body protein 2A
|
|
chr2_+_43864405
|
1.317
|
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr4_+_119512890
|
1.300
|
|
LOC729218
|
uncharacterized LOC729218
|
|
chr12_-_57914287
|
1.293
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr5_+_137673223
|
1.285
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr10_-_31320772
|
1.283
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr8_+_56686179
|
1.277
|
|
|
|
|
chr19_-_18392436
|
1.258
|
|
JUND
|
jun D proto-oncogene
|
|
chr10_+_5566923
|
1.255
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr2_+_228678556
|
1.251
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr5_+_137801166
|
1.251
|
|
EGR1
|
early growth response 1
|
|
chr2_+_43864433
|
1.248
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr2_-_86850935
|
1.246
|
|
RNF103
|
ring finger protein 103
|
|
chr12_+_132628978
|
1.239
|
NM_024078
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr17_-_8055684
|
1.232
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr19_-_18392413
|
1.224
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr3_+_158288952
|
1.218
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr17_-_8059637
|
1.215
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr15_-_80215663
|
1.209
|
NM_001100880
NM_001199757
|
ST20
|
suppressor of tumorigenicity 20
|
|
chr4_+_88571453
|
1.208
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr12_-_49504646
|
1.206
|
NM_018113
|
LMBR1L
|
limb region 1 homolog (mouse)-like
|
|
chr9_-_33402505
|
1.199
|
NM_001170
|
AQP7
|
aquaporin 7
|
|
chr2_+_149633173
|
1.194
|
|
|
|
|
chr3_-_50374749
|
1.194
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr17_-_13505217
|
1.192
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr7_-_140624280
|
1.185
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr20_+_32254303
|
1.182
|
NM_001024675
|
ACTL10
|
actin-like 10
|
|
chr3_-_47933997
|
1.181
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr16_-_54320367
|
1.172
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr6_+_31783280
|
1.171
|
NM_005345
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr9_+_130830447
|
1.168
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr2_+_37571752
|
1.168
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr9_+_130830595
|
1.166
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr19_+_55795766
|
1.165
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr19_+_13858687
|
1.164
|
NM_030818
|
CCDC130
|
coiled-coil domain containing 130
|
|
chr5_+_137801178
|
1.162
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr6_-_88411872
|
1.159
|
|
AKIRIN2
|
akirin 2
|
|
chr11_+_111749947
|
1.157
|
NM_022761
|
C11orf1
|
chromosome 11 open reading frame 1
|
|
chr4_-_89618928
|
1.154
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr5_-_175964824
|
1.153
|
|
RNF44
|
ring finger protein 44
|
|
chr7_+_20370324
|
1.153
|
|
ITGB8
|
integrin, beta 8
|
|
chr11_+_66610882
|
1.153
|
NM_001032279
NM_005133
|
RCE1
|
RCE1 homolog, prenyl protein protease (S. cerevisiae)
|
|
chr5_-_16617085
|
1.150
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr3_-_133614421
|
1.150
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr4_+_103422479
|
1.148
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr9_+_35490018
|
1.146
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr18_+_77439798
|
1.137
|
NM_004715
NM_048368
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr2_+_70142236
|
1.136
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr7_+_123295860
|
1.136
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr11_+_18343794
|
1.135
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr9_+_12693385
|
1.131
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr1_+_45266048
|
1.130
|
|
PLK3
|
polo-like kinase 3
|
|
chr4_-_103422436
|
1.128
|
|
|
|
|
chr21_+_47546427
|
1.127
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr2_-_86850959
|
1.126
|
NM_001198951
NM_001198952
NM_005667
|
RNF103
|
ring finger protein 103
|
|
chr17_-_62833264
|
1.122
|
|
PLEKHM1P
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene
|
|
chr17_-_8151373
|
1.119
|
NM_025099
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr3_+_150125983
|
1.116
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr8_-_101322245
|
1.106
|
NM_183419
|
RNF19A
|
ring finger protein 19A
|
|
chr4_-_2010685
|
1.097
|
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr6_+_31783314
|
1.097
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr2_+_33172368
|
1.096
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr13_+_97928196
|
1.094
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_+_62475165
|
1.087
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr19_-_42759151
|
1.073
|
|
ERF
|
Ets2 repressor factor
|
|
chr17_-_8151353
|
1.058
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr5_-_54468833
|
1.058
|
NM_001145734
NM_001170402
NM_152623
|
CDC20B
|
cell division cycle 20 homolog B (S. cerevisiae)
|
|
chr1_+_157963310
|
1.057
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr10_-_18940515
|
1.057
|
NM_182543
|
NSUN6
|
NOP2/Sun domain family, member 6
|
|
chr5_-_145483877
|
1.056
|
NM_001029869
|
PLAC8L1
|
PLAC8-like 1
|
|
chr11_-_69519105
|
1.053
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr18_+_77439843
|
1.048
|
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr17_+_42385780
|
1.042
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr2_+_10186284
|
1.041
|
|
|
|
|
chrX_-_52987610
|
1.040
|
NM_001242491
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr9_+_35489989
|
1.040
|
NM_014806
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr4_-_2010702
|
1.039
|
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chrX_+_84498993
|
1.036
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr5_-_178054001
|
1.034
|
NM_020666
|
CLK4
|
CDC-like kinase 4
|
|
chr21_-_44846927
|
1.030
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr1_-_1284755
|
1.028
|
|
DVL1
|
dishevelled, dsh homolog 1 (Drosophila)
|
|
chr17_-_43568081
|
1.024
|
|
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1
|
|
chr10_+_63808968
|
1.020
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr6_-_88411948
|
1.018
|
|
AKIRIN2
|
akirin 2
|
|
chr16_-_88772732
|
1.005
|
NM_001171815
|
RNF166
|
ring finger protein 166
|