|
chr11_-_1606480
|
7.359
|
NM_001005922
|
KRTAP5-1
|
keratin associated protein 5-1
|
|
chr16_-_67978422
|
6.786
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr5_+_137801178
|
6.568
|
NM_001964
|
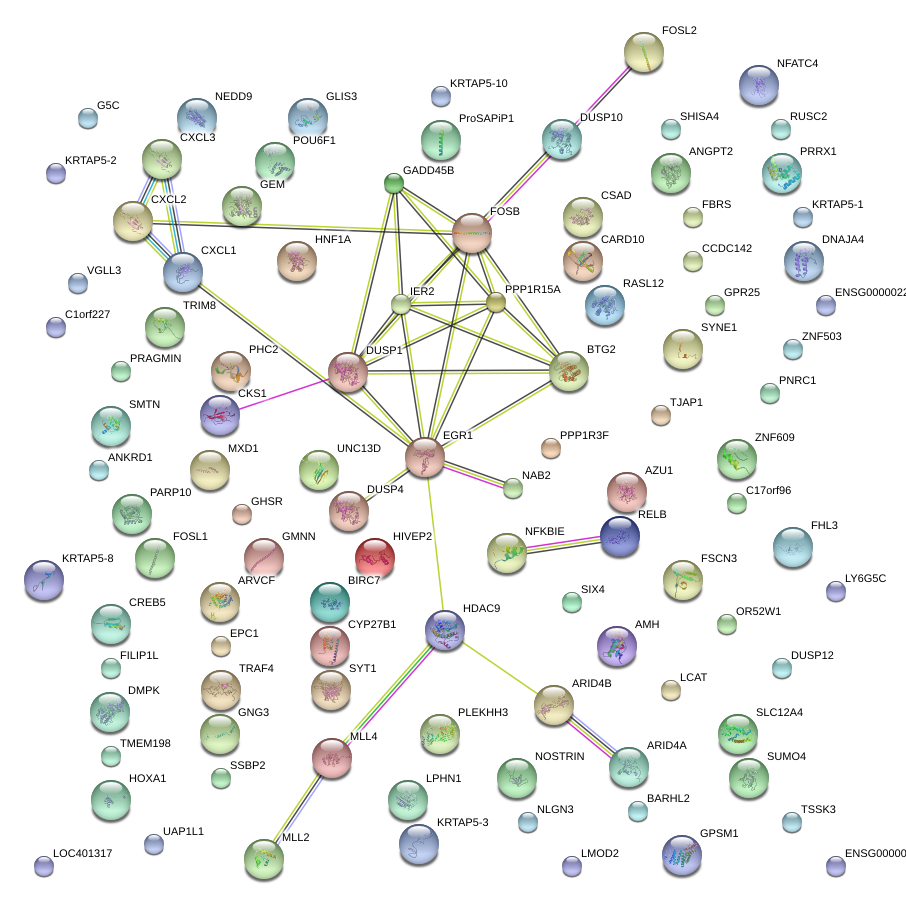
EGR1
|
early growth response 1
|
|
chr5_+_137801166
|
6.279
|
|
EGR1
|
early growth response 1
|
|
chr11_+_62475113
|
6.028
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr1_+_32827797
|
4.883
|
NM_052841
|
TSSK3
|
testis-specific serine kinase 3
|
|
chr6_+_149721494
|
4.748
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr11_+_62475165
|
4.692
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr7_+_28448970
|
4.587
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr22_+_31489343
|
4.468
|
|
SMTN
|
smoothelin
|
|
chr19_+_2249112
|
4.075
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr1_-_38471170
|
3.909
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr6_-_11382501
|
3.890
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_+_170633312
|
3.871
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr1_-_32827843
|
3.835
|
NM_001167676
|
LOC100128071
|
uncharacterized LOC100128071
|
|
chr3_-_169482847
|
3.814
|
|
TERC
|
telomerase RNA component
|
|
chr7_+_123295860
|
3.801
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr4_-_74964836
|
3.763
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr7_+_127233688
|
3.570
|
NM_020369
|
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus)
|
|
chr13_+_97999289
|
3.456
|
|
|
|
|
chr12_+_121416294
|
3.409
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr20_+_61867234
|
3.408
|
NM_022161
NM_139317
|
BIRC7
|
baculoviral IAP repeat containing 7
|
|
chr16_-_67977956
|
3.327
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr19_+_36208917
|
3.307
|
NM_014727
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr1_+_200842082
|
3.300
|
NM_005298
|
GPR25
|
G protein-coupled receptor 25
|
|
chr22_+_31487315
|
3.278
|
|
SMTN
|
smoothelin
|
|
chr6_-_152702450
|
3.242
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr1_-_91182777
|
3.227
|
NM_020063
|
BARHL2
|
BarH-like homeobox 2
|
|
chr12_+_57482886
|
3.224
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr12_+_57482922
|
3.223
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr1_-_213020947
|
3.187
|
NM_001024601
|
C1orf227
|
chromosome 1 open reading frame 227
|
|
chr19_-_14260728
|
3.169
|
|
LPHN1
|
latrophilin 1
|
|
chr19_-_46285752
|
3.157
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr12_+_121416548
|
3.133
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr7_+_18535351
|
3.125
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr19_+_13263890
|
3.123
|
|
IER2
|
immediate early response 2
|
|
chrX_+_49126299
|
3.100
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr4_-_74904329
|
3.002
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr19_-_46283281
|
2.962
|
|
|
|
|
chr15_-_65360297
|
2.892
|
NM_016563
|
RASL12
|
RAS-like, family 12
|
|
chr19_+_2476122
|
2.855
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr5_-_172198160
|
2.843
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr12_-_58159386
|
2.824
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr6_-_44233244
|
2.813
|
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chrX_+_49126480
|
2.805
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr16_+_30675777
|
2.801
|
NM_001105079
|
FBRS
|
fibrosin
|
|
chr1_-_33841142
|
2.791
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr8_-_6420454
|
2.791
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr2_-_74709965
|
2.768
|
|
CCDC142
|
coiled-coil domain containing 142
|
|
chr14_+_58765209
|
2.753
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr6_+_43457214
|
2.745
|
NM_001146017
|
TJAP1
|
tight junction associated protein 1 (peripheral)
|
|
chr10_-_32636134
|
2.739
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr3_-_87040268
|
2.730
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr1_-_221915393
|
2.695
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr17_+_27071008
|
2.694
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr2_+_70142236
|
2.675
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr4_-_74964996
|
2.668
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr6_+_89790464
|
2.619
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr8_-_29206321
|
2.614
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr1_+_201857997
|
2.612
|
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr1_+_203274646
|
2.607
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr11_-_65667809
|
2.582
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_+_49376488
|
2.576
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr2_+_28616360
|
2.565
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_-_27135524
|
2.533
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr15_+_64791618
|
2.531
|
NM_015042
|
ZNF609
|
zinc finger protein 609
|
|
chrX_+_70364680
|
2.525
|
NM_001166660
NM_018977
NM_181303
|
NLGN3
|
neuroligin 3
|
|
chr17_-_40828441
|
2.514
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr10_-_32636084
|
2.506
|
NM_025209
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr1_-_221915458
|
2.504
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr22_-_20004251
|
2.498
|
NM_001670
|
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome
|
|
chr3_-_172166202
|
2.489
|
NM_004122
NM_198407
|
GHSR
|
growth hormone secretagogue receptor
|
|
chrX_+_49126326
|
2.483
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr7_+_28475233
|
2.477
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_+_35538628
|
2.475
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_64498643
|
2.472
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr19_-_46285746
|
2.466
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr9_+_35538890
|
2.466
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr14_+_58765275
|
2.464
|
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr2_-_74710356
|
2.456
|
NM_032779
|
CCDC142
|
coiled-coil domain containing 142
|
|
chr6_+_89790412
|
2.451
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr17_-_36831156
|
2.413
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr5_-_81046858
|
2.394
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr10_-_92681025
|
2.369
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr8_-_8239188
|
2.363
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr1_-_221915426
|
2.359
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr12_-_53575121
|
2.344
|
|
CSAD
|
cysteine sulfinic acid decarboxylase
|
|
chr10_+_104404195
|
2.343
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr9_-_4299574
|
2.330
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr19_+_45971249
|
2.297
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr11_-_1619481
|
2.294
|
NM_001004325
|
KRTAP5-2
|
keratin associated protein 5-2
|
|
chr10_-_92680784
|
2.293
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr2_+_220408742
|
2.279
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr19_+_45504686
|
2.278
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr19_+_7660698
|
2.275
|
NM_001080429
NM_020902
|
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3
|
|
chr20_-_3149070
|
2.274
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr5_-_81046943
|
2.265
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr11_+_6220375
|
2.264
|
NM_001005178
|
OR52W1
|
olfactory receptor, family 52, subfamily W, member 1
|
|
chr3_-_99833348
|
2.257
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr19_+_9271367
|
2.199
|
|
|
|
|
chr12_-_51591949
|
2.190
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr9_+_139972195
|
2.183
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr14_+_24837225
|
2.169
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr19_+_827821
|
2.155
|
NM_001700
|
AZU1
|
azurocidin 1
|
|
chr8_-_95274540
|
2.138
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr4_+_74735107
|
2.115
|
NM_001511
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr8_-_145060594
|
2.113
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr14_-_61190458
|
2.100
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr9_+_139247508
|
2.099
|
NM_001145639
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr2_+_169659099
|
2.088
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr22_-_37915209
|
2.082
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr8_-_145060563
|
2.078
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr7_-_151071696
|
2.073
|
|
|
|
|
chr6_-_143266283
|
2.066
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr10_-_77159851
|
2.053
|
|
ZNF503
|
zinc finger protein 503
|
|
chr12_+_79258517
|
2.052
|
|
SYT1
|
synaptotagmin I
|
|
chr15_+_78556485
|
2.049
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr6_-_31648140
|
2.047
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr19_+_17830302
|
2.040
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr6_+_108881010
|
2.036
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr3_-_42814734
|
2.032
|
NM_144719
|
CCDC13
|
coiled-coil domain containing 13
|
|
chrX_+_95939661
|
2.028
|
NM_006729
NM_007309
|
DIAPH2
|
diaphanous homolog 2 (Drosophila)
|
|
chr6_-_33266297
|
2.025
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr6_-_30652434
|
2.021
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr22_-_40859423
|
2.019
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr11_-_117100551
|
2.011
|
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr5_-_176936857
|
2.005
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr12_+_57482674
|
2.004
|
NM_005967
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr6_-_41703345
|
1.996
|
|
TFEB
|
transcription factor EB
|
|
chr13_+_24144402
|
1.991
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr7_+_100081549
|
1.989
|
NM_173564
|
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1
|
|
chr9_+_130860816
|
1.984
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr7_-_141673572
|
1.984
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr19_+_36209125
|
1.981
|
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr17_-_7232455
|
1.977
|
NM_001005408
NM_032442
|
NEURL4
|
neuralized homolog 4 (Drosophila)
|
|
chr9_+_130860769
|
1.972
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr3_-_133614421
|
1.955
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr12_-_58160823
|
1.955
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr11_-_1507810
|
1.954
|
NM_001172223
|
MOB2
|
MOB kinase activator 2
|
|
chr3_+_50306578
|
1.953
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr11_+_5968576
|
1.951
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr17_-_1548992
|
1.939
|
NM_003693
NM_145350
NM_145352
|
SCARF1
|
scavenger receptor class F, member 1
|
|
chr7_-_100882100
|
1.933
|
NM_001185080
|
CLDN15
|
claudin 15
|
|
chr12_+_79258588
|
1.933
|
|
SYT1
|
synaptotagmin I
|
|
chr8_-_95274520
|
1.928
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr19_+_11649578
|
1.920
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chrX_-_103087106
|
1.920
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr12_+_79258448
|
1.917
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr5_+_55149149
|
1.916
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr8_-_8175860
|
1.898
|
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr1_-_221910801
|
1.893
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr17_+_37821598
|
1.893
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr9_-_123689172
|
1.891
|
NM_005658
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr22_-_39268169
|
1.881
|
|
CBX6
|
chromobox homolog 6
|
|
chrX_+_49047887
|
1.875
|
|
|
|
|
chr22_-_39268214
|
1.873
|
|
CBX6
|
chromobox homolog 6
|
|
chr1_-_114354906
|
1.872
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr4_-_185726865
|
1.867
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr4_+_3443659
|
1.864
|
|
HGFAC
|
HGF activator
|
|
chr2_+_220408352
|
1.856
|
|
TMEM198
|
transmembrane protein 198
|
|
chr17_+_7743233
|
1.854
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr10_-_32635842
|
1.845
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr3_+_50306305
|
1.845
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr19_+_544026
|
1.844
|
NM_005317
|
GZMM
|
granzyme M (lymphocyte met-ase 1)
|
|
chr18_+_29598787
|
1.837
|
|
RNF125
|
ring finger protein 125
|
|
chr16_-_75301950
|
1.832
|
NM_001170715
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr1_-_234743488
|
1.831
|
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chr12_-_112630525
|
1.825
|
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr14_+_55168831
|
1.822
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr14_-_24020766
|
1.816
|
NM_033400
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr8_+_128806720
|
1.811
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr7_+_107110501
|
1.810
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr22_+_31480981
|
1.808
|
NM_001207018
|
SMTN
|
smoothelin
|
|
chr15_+_89182038
|
1.802
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr2_+_29038862
|
1.800
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr16_+_69984809
|
1.796
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr19_-_40724272
|
1.793
|
NM_152479
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr4_+_108745718
|
1.790
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr1_+_36554452
|
1.785
|
NM_017825
|
ADPRHL2
|
ADP-ribosylhydrolase like 2
|
|
chr14_+_103592663
|
1.782
|
NM_006291
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr22_-_37640287
|
1.781
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr3_-_114478117
|
1.777
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_-_33267128
|
1.774
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr3_+_119421865
|
1.773
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr15_+_85359910
|
1.771
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr1_-_45272956
|
1.763
|
NM_001013632
|
TCTEX1D4
|
Tctex1 domain containing 4
|
|
chr3_-_87040246
|
1.762
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr9_+_34179058
|
1.751
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr9_-_34662629
|
1.742
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr17_+_18855477
|
1.726
|
NM_001042450
NM_152351
|
SLC5A10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10
|
|
chr1_+_177140530
|
1.719
|
NM_021165
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr16_-_58033761
|
1.711
|
NM_020807
|
ZNF319
|
zinc finger protein 319
|
|
chr17_-_1395950
|
1.711
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr12_-_54778298
|
1.711
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr17_-_37353731
|
1.706
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr1_+_63788729
|
1.706
|
NM_012183
|
FOXD3
|
forkhead box D3
|
|
chr1_-_114354864
|
1.705
|
|
RSBN1
|
round spermatid basic protein 1
|
|
chr7_+_18330044
|
1.701
|
|
HDAC9
|
histone deacetylase 9
|
|
chr17_+_26698666
|
1.700
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr19_-_13947101
|
1.694
|
|
LOC284454
|
uncharacterized LOC284454
|