|
chr1_+_995109
|
4.964
|
|
LOC100288175
|
uncharacterized LOC100288175
|
|
chr20_-_3388219
|
4.762
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr21_-_37528578
|
4.059
|
|
LOC100506428
|
uncharacterized LOC100506428
|
|
chr19_+_45504686
|
3.853
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr13_-_110438896
|
3.529
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr9_+_139972195
|
3.189
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr20_-_3388586
|
3.113
|
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr20_-_56884488
|
3.082
|
|
PPP4R1L
|
protein phosphatase 4, regulatory subunit 1-like
|
|
chr9_-_110251754
|
3.081
|
|
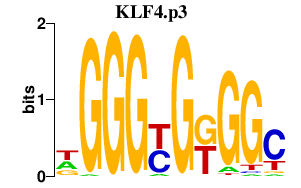
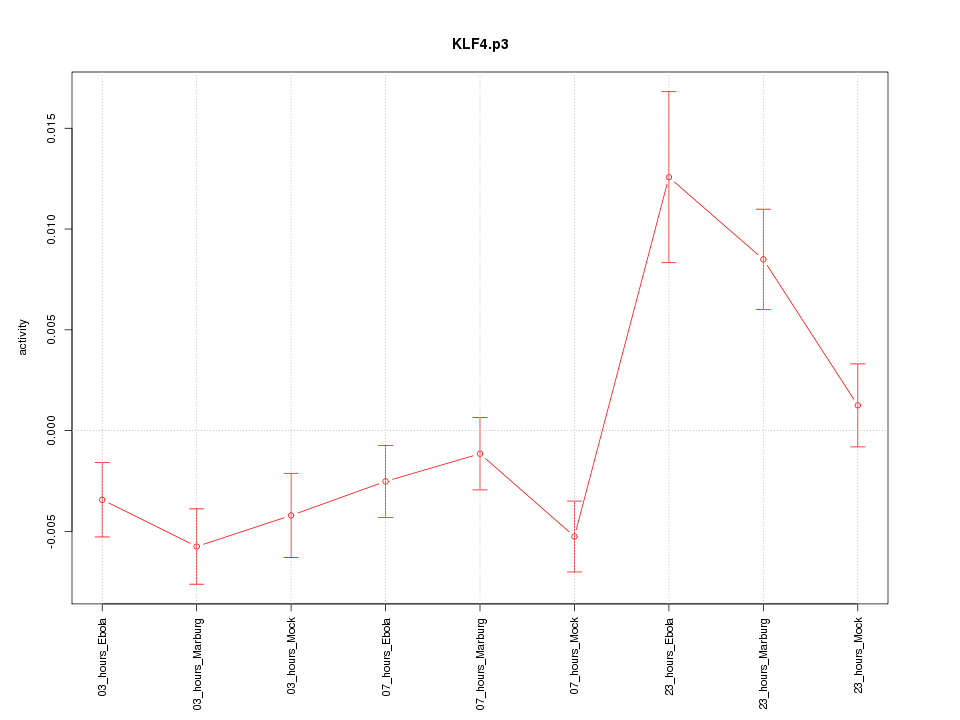
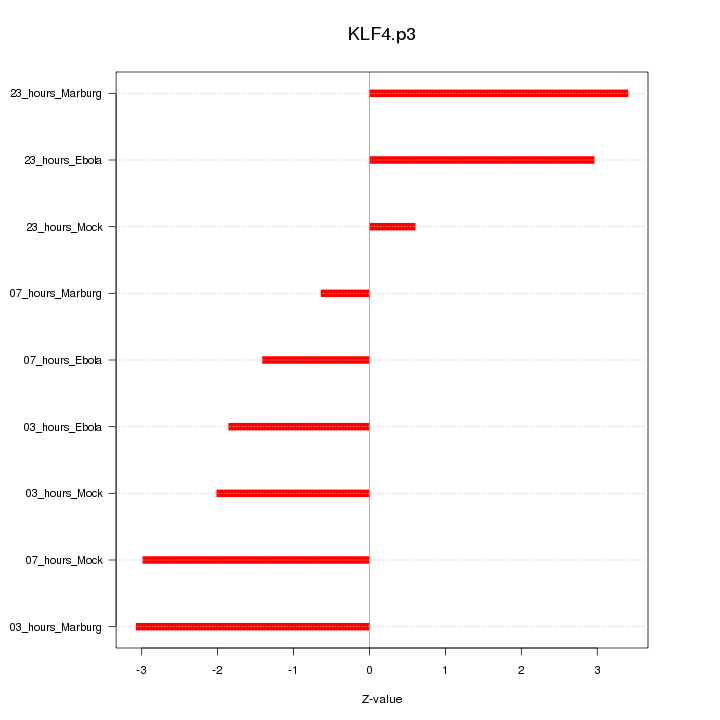
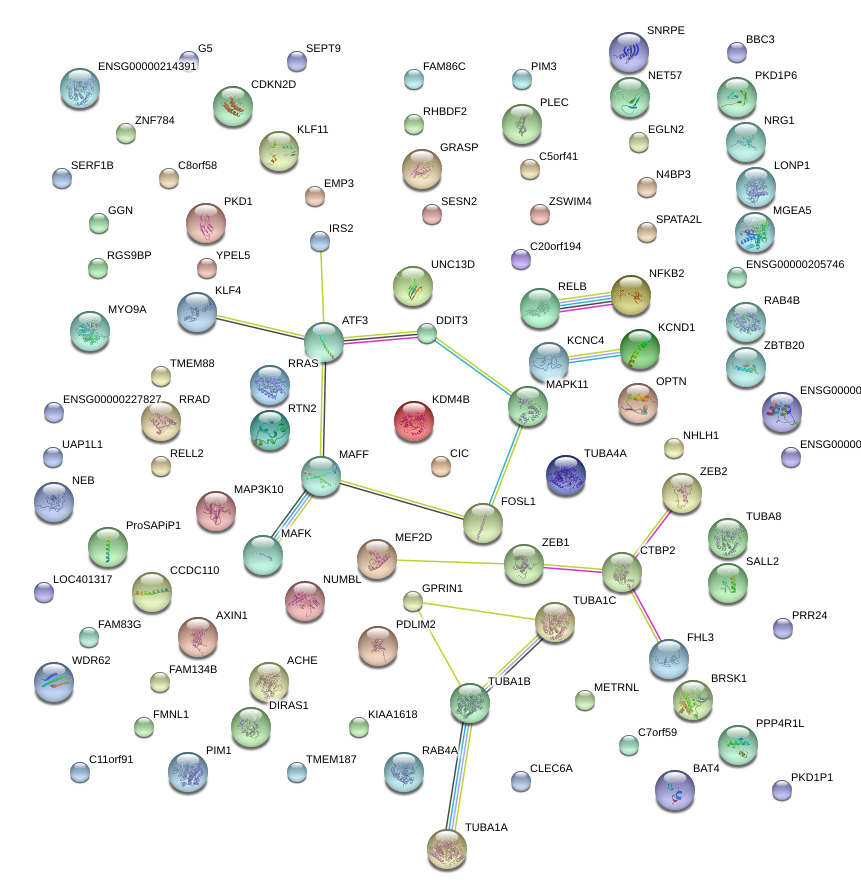
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr5_+_172484353
|
3.066
|
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr7_+_64498643
|
2.974
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr2_+_10183630
|
2.920
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr22_+_50354428
|
2.917
|
|
PIM3
|
pim-3 oncogene
|
|
chr19_-_47734215
|
2.903
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr6_+_37137882
|
2.900
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr3_-_114866090
|
2.849
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr19_+_55795766
|
2.754
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr19_+_41284161
|
2.729
|
|
RAB4B-EGLN2
RAB4B
|
RAB4B-EGLN2 readthrough
RAB4B, member RAS oncogene family
|
|
chr9_+_139971938
|
2.721
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr17_+_43299277
|
2.626
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr15_-_72410108
|
2.616
|
|
MYO9A
|
myosin IXA
|
|
chr22_-_50708720
|
2.539
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr10_+_104155431
|
2.501
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr7_-_100493481
|
2.495
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr19_+_33166312
|
2.478
|
NM_207391
|
RGS9BP
|
regulator of G protein signaling 9 binding protein
|
|
chr19_-_41196419
|
2.456
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr19_-_38878663
|
2.438
|
NM_152657
|
GGN
|
gametogenetin
|
|
chr5_+_177540465
|
2.431
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr1_+_212782103
|
2.427
|
|
ATF3
|
activating transcription factor 3
|
|
chr3_-_129830273
|
2.402
|
|
|
|
|
chr19_-_10679328
|
2.337
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr7_+_1570321
|
2.329
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr12_+_52400725
|
2.309
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr5_+_172483285
|
2.308
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr12_-_57914287
|
2.270
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr3_-_129830320
|
2.265
|
|
|
|
|
chr7_+_28448970
|
2.246
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr1_+_28585959
|
2.218
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr20_-_3154169
|
2.215
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr15_-_72410226
|
2.198
|
NM_006901
|
MYO9A
|
myosin IXA
|
|
chr10_+_13142055
|
2.158
|
NM_001008211
NM_001008212
NM_001008213
NM_021980
|
OPTN
|
optineurin
|
|
chr22_+_50354139
|
2.151
|
NM_001001852
|
PIM3
|
pim-3 oncogene
|
|
chr17_+_81037504
|
2.123
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr1_-_38471170
|
2.120
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr1_+_160336856
|
2.112
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr16_-_2185824
|
2.100
|
NM_000296
NM_001009944
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chrX_-_48828250
|
2.078
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr19_+_42788605
|
2.073
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr8_+_8086103
|
2.026
|
|
FLJ10661
|
family with sequence similarity 86, member A pseudogene
|
|
chr2_-_145274916
|
2.023
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr8_+_22457098
|
2.023
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr19_+_47778099
|
2.015
|
NM_178511
|
PRR24
|
proline rich 24
|
|
chr17_-_74497406
|
1.998
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr12_-_49582296
|
1.996
|
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr19_-_50143350
|
1.977
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr19_+_4969123
|
1.967
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr19_-_46000159
|
1.961
|
NM_005619
NM_206900
|
RTN2
|
reticulon 2
|
|
chr11_-_65667809
|
1.929
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_+_40697516
|
1.920
|
NM_002446
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10
|
|
chr7_+_100081549
|
1.915
|
NM_173564
|
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1
|
|
chr4_-_186392847
|
1.903
|
NM_001145411
NM_152775
|
CCDC110
|
coiled-coil domain containing 110
|
|
chr1_-_1475739
|
1.892
|
NM_001114748
|
TMEM240
|
transmembrane protein 240
|
|
chr16_-_402617
|
1.889
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr1_+_110753313
|
1.858
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr5_-_176037091
|
1.856
|
NM_052899
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr19_+_41305100
|
1.847
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_+_48828820
|
1.844
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr8_+_32405727
|
1.843
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr5_+_141016516
|
1.836
|
NM_001130029
NM_173828
|
RELL2
|
RELT-like 2
|
|
chr16_-_89768094
|
1.803
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr19_+_41305041
|
1.799
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chrX_+_153237990
|
1.795
|
NM_003492
|
TMEM187
|
transmembrane protein 187
|
|
chr11_-_33722285
|
1.781
|
NM_001166692
|
C11orf91
|
chromosome 11 open reading frame 91
|
|
chr17_+_78234626
|
1.777
|
NM_020914
NM_020954
|
RNF213
|
ring finger protein 213
|
|
chr8_-_145013628
|
1.773
|
NM_201384
|
PLEC
|
plectin
|
|
chr19_-_56135931
|
1.769
|
|
ZNF784
|
zinc finger protein 784
|
|
chr17_-_18907988
|
1.764
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr10_-_126849556
|
1.743
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr19_-_56135934
|
1.698
|
NM_203374
|
ZNF784
|
zinc finger protein 784
|
|
chr17_+_7758380
|
1.694
|
NM_203411
|
TMEM88
|
transmembrane protein 88
|
|
chr1_+_212781969
|
1.688
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr2_+_30369868
|
1.687
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr5_-_16617085
|
1.684
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr6_-_31633587
|
1.681
|
NM_001199238
NM_001199239
|
GPANK1
|
G patch domain and ankyrin repeats 1
|
|
chr9_-_35115844
|
1.680
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr1_-_156470538
|
1.679
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr19_-_2721337
|
1.673
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr16_-_66959418
|
1.664
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr19_-_10679620
|
1.660
|
NM_001800
NM_079421
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr22_+_38597938
|
1.654
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr2_+_149633173
|
1.638
|
|
|
|
|
chr19_+_13906207
|
1.625
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr1_+_45266048
|
1.623
|
|
PLK3
|
polo-like kinase 3
|
|
chr3_-_109035239
|
1.623
|
NM_138815
|
DPPA2
|
developmental pluripotency associated 2
|
|
chr19_+_4969126
|
1.618
|
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr12_+_120933916
|
1.612
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr19_-_19738973
|
1.612
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr9_-_139890984
|
1.611
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr2_+_28615668
|
1.608
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr22_+_38599026
|
1.606
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr22_+_38598065
|
1.603
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr22_+_31477231
|
1.599
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr1_-_156470506
|
1.592
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr15_-_60884615
|
1.591
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_-_110252045
|
1.591
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr6_-_44233499
|
1.583
|
NM_004556
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr15_+_90931470
|
1.580
|
NM_003870
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr8_+_144329105
|
1.579
|
NM_173832
|
ZFP41
|
zinc finger protein 41 homolog (mouse)
|
|
chr19_-_1237840
|
1.579
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr14_-_71275732
|
1.578
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr2_+_30369749
|
1.571
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr19_+_47759802
|
1.570
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr14_-_105767328
|
1.566
|
NM_001242788
NM_001242790
NM_001519
|
BRF1
|
BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae)
|
|
chr3_-_132441302
|
1.551
|
NM_153240
|
NPHP3-ACAD11
NPHP3
|
NPHP3-ACAD11 readthrough
nephronophthisis 3 (adolescent)
|
|
chr12_+_58148775
|
1.546
|
NM_138396
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9
|
|
chr6_-_41702734
|
1.545
|
NM_007162
|
TFEB
|
transcription factor EB
|
|
chr12_+_57482922
|
1.543
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr12_+_120933832
|
1.537
|
NM_001037495
NM_003746
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr19_-_18632886
|
1.527
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr6_-_44233244
|
1.512
|
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr11_-_1593149
|
1.511
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr8_-_145050896
|
1.499
|
NM_000445
|
PLEC
|
plectin
|
|
chr12_-_100378431
|
1.481
|
NM_152788
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr6_-_33266735
|
1.480
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr19_+_7660698
|
1.480
|
NM_001080429
NM_020902
|
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3
|
|
chr12_+_106696508
|
1.476
|
NM_152772
|
TCP11L2
|
t-complex 11 (mouse)-like 2
|
|
chr12_-_49582800
|
1.476
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr20_+_388717
|
1.472
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr1_+_35331533
|
1.472
|
|
|
|
|
chr6_-_33285718
|
1.471
|
NM_001145338
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr17_-_49198084
|
1.471
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr17_-_72968790
|
1.469
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr16_+_66638652
|
1.468
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr4_-_82392971
|
1.467
|
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr19_-_46283281
|
1.463
|
|
|
|
|
chr1_-_212873197
|
1.462
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr19_+_55795533
|
1.461
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr7_-_2354050
|
1.459
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr19_-_36523540
|
1.451
|
NM_001199570
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr8_-_29207795
|
1.443
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr19_+_49617631
|
1.438
|
|
LIN7B
|
lin-7 homolog B (C. elegans)
|
|
chr8_+_144680073
|
1.438
|
NM_032862
|
TIGD5
|
tigger transposable element derived 5
|
|
chr12_+_58120029
|
1.425
|
|
LOC100130776
|
uncharacterized LOC100130776
|
|
chr19_-_4065467
|
1.423
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr12_-_112819755
|
1.422
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr6_-_33266700
|
1.418
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr9_+_139221931
|
1.417
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr19_-_18632857
|
1.412
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr6_+_138188352
|
1.411
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_-_6321034
|
1.411
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr8_+_144328990
|
1.409
|
|
ZFP41
|
zinc finger protein 41 homolog (mouse)
|
|
chr11_-_65667860
|
1.409
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr7_+_99775521
|
1.409
|
NM_012447
|
STAG3
|
stromal antigen 3
|
|
chr19_-_11373117
|
1.407
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr5_+_141016567
|
1.406
|
|
RELL2
|
RELT-like 2
|
|
chr11_+_46299161
|
1.402
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr7_-_2354010
|
1.401
|
|
SNX8
|
sorting nexin 8
|
|
chr17_-_17109601
|
1.400
|
|
PLD6
|
phospholipase D family, member 6
|
|
chr2_+_10184371
|
1.394
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr2_+_23608297
|
1.394
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr12_+_57482674
|
1.390
|
NM_005967
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr9_-_110251308
|
1.390
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr19_-_1111972
|
1.389
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr22_+_51039178
|
1.389
|
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr8_-_145016691
|
1.389
|
NM_201383
|
PLEC
|
plectin
|
|
chr17_-_17109630
|
1.387
|
NM_178836
|
PLD6
|
phospholipase D family, member 6
|
|
chr19_+_49622480
|
1.386
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr14_+_24838301
|
1.379
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr5_+_179234002
|
1.377
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr9_-_35732389
|
1.376
|
NM_006289
|
TLN1
|
talin 1
|
|
chr6_+_108882104
|
1.372
|
|
FOXO3
|
forkhead box O3
|
|
chr19_+_41305333
|
1.367
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr11_+_77899857
|
1.366
|
NM_020798
|
USP35
|
ubiquitin specific peptidase 35
|
|
chr6_+_17281773
|
1.361
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_-_17699705
|
1.359
|
NM_001099218
|
RAD51AP2
|
RAD51 associated protein 2
|
|
chr16_+_66638632
|
1.354
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr12_+_57482886
|
1.354
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr1_+_45266035
|
1.350
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr15_+_90931489
|
1.347
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr16_-_30381487
|
1.344
|
NM_015527
|
TBC1D10B
|
TBC1 domain family, member 10B
|
|
chr8_-_144650998
|
1.343
|
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr6_-_33285493
|
1.340
|
NM_005453
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr17_-_41910439
|
1.340
|
NM_001932
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr14_+_75745480
|
1.337
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr16_+_2039945
|
1.334
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr12_+_111843743
|
1.327
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr9_+_99212379
|
1.325
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr19_-_40971623
|
1.324
|
NM_000713
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH))
|
|
chr15_-_61521416
|
1.322
|
|
RORA
|
RAR-related orphan receptor A
|
|
chr7_-_4923246
|
1.319
|
NM_018059
|
RADIL
|
Ras association and DIL domains
|
|
chr1_-_38273852
|
1.315
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr1_+_233463513
|
1.315
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chr16_+_1203240
|
1.314
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr11_-_64684606
|
1.313
|
NM_015104
|
ATG2A
|
ATG2 autophagy related 2 homolog A (S. cerevisiae)
|
|
chr19_+_41284068
|
1.307
|
NM_016154
|
RAB4B
|
RAB4B, member RAS oncogene family
|
|
chr7_+_73082157
|
1.304
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr1_+_37940129
|
1.302
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr13_+_52027435
|
1.301
|
|
|
|
|
chr14_+_105331581
|
1.299
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr15_+_90931449
|
1.296
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|