|
chr9_-_110251754
|
2.300
|
|
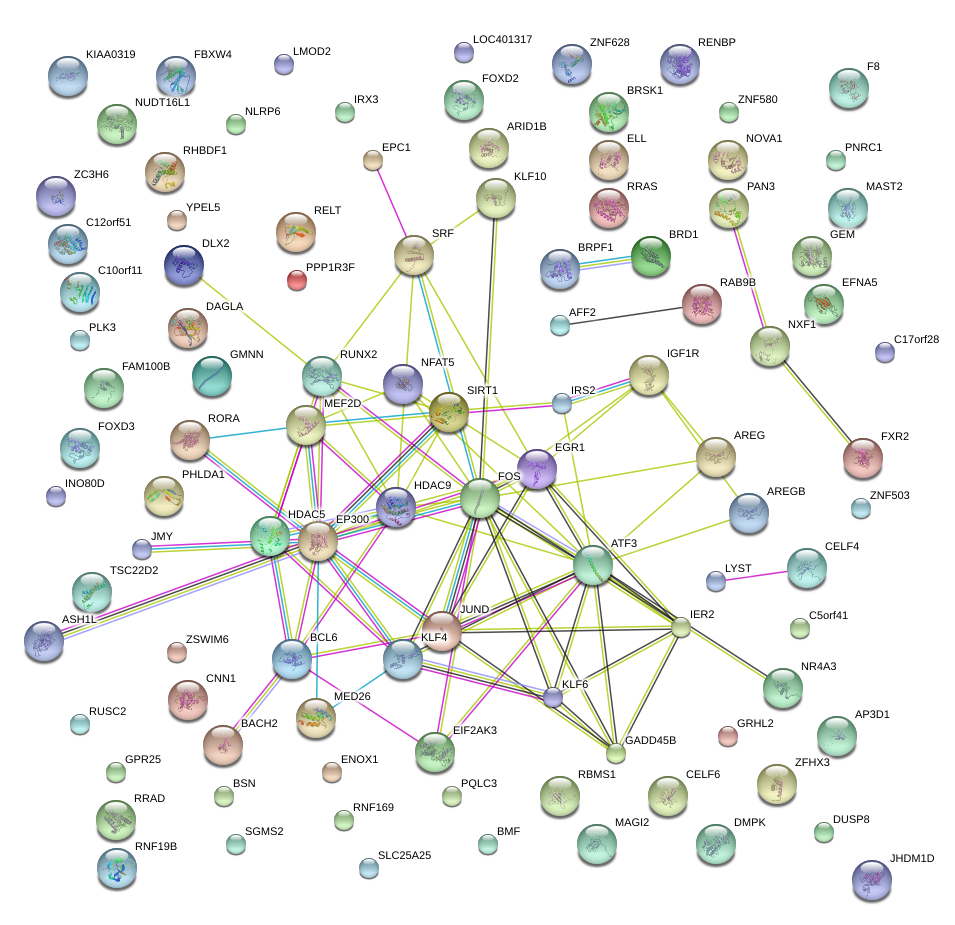
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr6_-_91006460
|
2.043
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr19_+_11649578
|
2.028
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr12_-_76425207
|
1.987
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr3_+_49591897
|
1.981
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr5_+_137801178
|
1.930
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr3_-_169482847
|
1.879
|
|
TERC
|
telomerase RNA component
|
|
chr17_-_7518124
|
1.802
|
NM_004860
|
FXR2
|
fragile X mental retardation, autosomal homolog 2
|
|
chr9_-_110251308
|
1.601
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chrX_+_147582133
|
1.588
|
NM_001169122
NM_001169123
NM_001169124
NM_001169125
NM_002025
|
AFF2
|
AF4/FMR2 family, member 2
|
|
chr19_+_2476122
|
1.569
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr17_+_56065408
|
1.523
|
|
|
|
|
chrX_+_49126480
|
1.497
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr8_-_95274540
|
1.399
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr9_+_35560424
|
1.386
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_123295860
|
1.379
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr1_+_63788729
|
1.379
|
NM_012183
|
FOXD3
|
forkhead box D3
|
|
chr6_-_91006580
|
1.358
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chrX_+_49126299
|
1.325
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr16_+_4743709
|
1.311
|
|
NUDT16L1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1
|
|
chr6_+_89791510
|
1.302
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr7_+_28448970
|
1.296
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr8_-_95274520
|
1.294
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr2_+_30369749
|
1.294
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr17_+_74261280
|
1.284
|
NM_182565
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr14_+_75745480
|
1.266
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr10_-_77161136
|
1.250
|
|
ZNF503
|
zinc finger protein 503
|
|
chr2_-_88926863
|
1.223
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr16_+_69600110
|
1.219
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chrX_+_49126326
|
1.216
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr19_+_13263890
|
1.183
|
|
IER2
|
immediate early response 2
|
|
chr19_-_18632857
|
1.173
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr16_+_4743693
|
1.167
|
NM_001193452
NM_032349
|
NUDT16L1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1
|
|
chr17_+_74261431
|
1.161
|
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr1_+_212781969
|
1.158
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr17_-_72968790
|
1.153
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr11_-_1593149
|
1.141
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr6_+_135818938
|
1.133
|
|
LINC00271
|
long intergenic non-protein coding RNA 271
|
|
chr5_-_107006585
|
1.129
|
NM_001962
|
EFNA5
|
ephrin-A5
|
|
chr2_-_206950895
|
1.127
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr2_-_172967014
|
1.122
|
|
DLX2
|
distal-less homeobox 2
|
|
chr5_+_172483285
|
1.107
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr2_+_113033184
|
1.103
|
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr17_-_72968750
|
1.102
|
|
|
|
|
chr11_+_74459900
|
1.095
|
NM_001098638
|
RNF169
|
ring finger protein 169
|
|
chr2_+_113033146
|
1.095
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr9_-_110252045
|
1.092
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_-_33430285
|
1.091
|
NM_001127361
NM_153341
|
RNF19B
|
ring finger protein 19B
|
|
chr5_+_137801166
|
1.090
|
|
EGR1
|
early growth response 1
|
|
chr1_+_212782103
|
1.088
|
|
ATF3
|
activating transcription factor 3
|
|
chr12_-_76425375
|
1.087
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr6_+_157099873
|
1.077
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr19_-_18632886
|
1.076
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr10_-_77160650
|
1.061
|
|
ZNF503
|
zinc finger protein 503
|
|
chr13_-_110438896
|
1.060
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr8_-_103667882
|
1.059
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr2_+_30369868
|
1.054
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr18_-_35145603
|
1.049
|
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr16_-_66959418
|
1.040
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr15_-_61521416
|
1.037
|
|
RORA
|
RAR-related orphan receptor A
|
|
chr16_-_54319966
|
1.027
|
|
IRX3
|
iroquois homeobox 3
|
|
chr9_+_102584136
|
1.022
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr19_+_55795533
|
1.017
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr2_+_30369800
|
1.015
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chrX_-_153210057
|
1.015
|
NM_002910
|
RENBP
|
renin binding protein
|
|
chr15_-_72612247
|
0.998
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr17_-_7517849
|
0.993
|
|
FXR2
|
fragile X mental retardation, autosomal homolog 2
|
|
chr7_+_18548899
|
0.988
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr10_-_77161424
|
0.987
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr13_-_44361032
|
0.985
|
NM_001242863
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr3_-_187463474
|
0.985
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr10_-_103454687
|
0.969
|
NM_022039
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chr19_+_55987698
|
0.966
|
NM_033113
|
ZNF628
|
zinc finger protein 628
|
|
chr2_-_161350304
|
0.964
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr7_-_79082870
|
0.961
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr11_-_62572827
|
0.960
|
NM_001081491
NM_006362
|
NXF1
|
nuclear RNA export factor 1
|
|
chr22_+_41488607
|
0.959
|
NM_001429
|
EP300
|
E1A binding protein p300
|
|
chr5_+_60628085
|
0.953
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr16_-_122573
|
0.948
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr1_-_236030219
|
0.941
|
NM_000081
|
LYST
|
lysosomal trafficking regulator
|
|
chr6_+_43139190
|
0.940
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr3_-_187463227
|
0.937
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr10_+_74451838
|
0.931
|
NM_138357
|
MCU
|
mitochondrial calcium uniporter
|
|
chr15_+_99191671
|
0.928
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr15_-_40401041
|
0.927
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr9_+_130854127
|
0.924
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr3_+_9773439
|
0.920
|
|
BRPF1
|
bromodomain and PHD finger containing, 1
|
|
chr16_-_73092533
|
0.914
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr19_-_16738986
|
0.913
|
NM_004831
|
MED26
|
mediator complex subunit 26
|
|
chr4_+_108745718
|
0.900
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr2_+_11295539
|
0.898
|
NM_152391
|
PQLC3
|
PQ loop repeat containing 3
|
|
chr1_+_200842082
|
0.894
|
NM_005298
|
GPR25
|
G protein-coupled receptor 25
|
|
chr5_+_78532338
|
0.894
|
|
|
|
|
chr19_+_56152281
|
0.893
|
NM_207115
|
ZNF580
|
zinc finger protein 580
|
|
chr10_+_77542518
|
0.892
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr13_+_28712379
|
0.874
|
NM_175854
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr7_-_139876718
|
0.868
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr2_-_172967627
|
0.864
|
|
DLX2
|
distal-less homeobox 2
|
|
chr19_+_45596406
|
0.863
|
NM_019121
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37
|
|
chr2_-_172967234
|
0.861
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr19_-_2151551
|
0.860
|
NM_001077523
NM_003938
|
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit
|
|
chrX_-_103087106
|
0.860
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr10_-_3827418
|
0.859
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr12_-_112819755
|
0.858
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr11_+_61447816
|
0.857
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr3_+_150125983
|
0.855
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr1_+_45266048
|
0.853
|
|
PLK3
|
polo-like kinase 3
|
|
chr5_+_78531848
|
0.842
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr10_+_69644938
|
0.842
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr15_-_61521478
|
0.838
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_-_156470506
|
0.837
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr19_-_18392048
|
0.837
|
|
JUND
|
jun D proto-oncogene
|
|
chr11_+_73087670
|
0.835
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr1_-_156470538
|
0.830
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr16_-_54318889
|
0.828
|
|
IRX3
|
iroquois homeobox 3
|
|
chr1_-_155532152
|
0.824
|
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr1_+_53099015
|
0.823
|
NM_001042693
|
FAM159A
|
family with sequence similarity 159, member A
|
|
chr6_+_45390284
|
0.821
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr14_-_27066959
|
0.818
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr4_+_75480628
|
0.817
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr10_-_32635842
|
0.817
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr1_+_46269283
|
0.812
|
NM_015112
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr19_-_46285752
|
0.807
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr17_-_42200957
|
0.806
|
|
HDAC5
|
histone deacetylase 5
|
|
chrX_-_154114431
|
0.805
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr1_+_16174358
|
0.803
|
NM_015001
|
SPEN
|
spen homolog, transcriptional regulator (Drosophila)
|
|
chr2_+_189156603
|
0.803
|
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr19_-_59069747
|
0.801
|
|
UBE2M
|
ubiquitin-conjugating enzyme E2M
|
|
chr6_+_149721494
|
0.800
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr19_+_55795766
|
0.797
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr5_-_57755787
|
0.796
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr1_+_177140530
|
0.796
|
NM_021165
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr2_+_189156653
|
0.793
|
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr17_-_27332888
|
0.793
|
NM_001098635
NM_178860
|
SEZ6
|
seizure related 6 homolog (mouse)
|
|
chr6_-_111804413
|
0.791
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr5_-_139725860
|
0.790
|
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr14_-_61190458
|
0.788
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr1_-_156470565
|
0.787
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr17_-_42201009
|
0.784
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr2_-_157189040
|
0.784
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr19_-_2151534
|
0.781
|
|
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit
|
|
chr7_+_18548963
|
0.781
|
|
HDAC9
|
histone deacetylase 9
|
|
chr1_-_1334648
|
0.778
|
NM_001039577
NM_030937
|
CCNL2
|
cyclin L2
|
|
chr3_-_114343052
|
0.777
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr4_+_15004297
|
0.777
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr12_-_76424474
|
0.776
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr6_+_12012723
|
0.775
|
NM_002114
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1
|
|
chr6_-_150039391
|
0.771
|
NM_004690
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr2_+_198318225
|
0.768
|
NM_025147
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae)
|
|
chr6_+_37787592
|
0.767
|
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr19_+_49375638
|
0.764
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr5_+_173315313
|
0.758
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr22_-_17602142
|
0.758
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr15_-_40212425
|
0.758
|
|
GPR176
|
G protein-coupled receptor 176
|
|
chr19_+_49375677
|
0.752
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr7_+_27135672
|
0.749
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr10_+_60272868
|
0.749
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr1_-_149908098
|
0.748
|
NM_181873
|
MTMR11
|
myotubularin related protein 11
|
|
chr17_-_18907988
|
0.743
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr7_-_103629962
|
0.740
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr19_+_45596159
|
0.735
|
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37
|
|
chr22_+_31489343
|
0.733
|
|
SMTN
|
smoothelin
|
|
chr1_-_155532562
|
0.730
|
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr9_-_80646150
|
0.728
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr4_-_74124456
|
0.724
|
NM_032217
NM_198889
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr4_-_89618928
|
0.723
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr15_-_65360297
|
0.716
|
NM_016563
|
RASL12
|
RAS-like, family 12
|
|
chr19_+_4791757
|
0.715
|
|
FEM1A
|
fem-1 homolog a (C. elegans)
|
|
chr10_+_69644708
|
0.712
|
|
SIRT1
|
sirtuin 1
|
|
chr7_-_100808832
|
0.712
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr19_+_49375679
|
0.711
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr5_-_11904127
|
0.710
|
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr8_-_8239188
|
0.709
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr7_+_128312345
|
0.706
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr3_-_114102488
|
0.703
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr19_+_12902295
|
0.702
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr7_+_138145179
|
0.700
|
|
TRIM24
|
tripartite motif containing 24
|
|
chr16_+_69599869
|
0.698
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr3_+_9773427
|
0.698
|
NM_001003694
NM_004634
|
BRPF1
|
bromodomain and PHD finger containing, 1
|
|
chr19_+_13135368
|
0.695
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_-_18392436
|
0.692
|
|
JUND
|
jun D proto-oncogene
|
|
chr19_+_13105970
|
0.691
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_+_50794591
|
0.689
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr6_-_150039159
|
0.688
|
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr11_+_65837785
|
0.688
|
NM_018026
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr3_-_134093235
|
0.688
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr7_-_99756275
|
0.685
|
NM_018275
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr4_+_103422553
|
0.684
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr3_+_51989329
|
0.681
|
NM_080865
|
GPR62
|
G protein-coupled receptor 62
|
|
chr9_-_3525721
|
0.678
|
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr7_+_64498643
|
0.677
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr1_-_59248287
|
0.677
|
|
JUN
|
jun proto-oncogene
|
|
chr19_-_40724272
|
0.671
|
NM_152479
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr7_+_6629658
|
0.670
|
NM_024067
|
C7orf26
|
chromosome 7 open reading frame 26
|
|
chr5_+_139060186
|
0.670
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr17_+_59477169
|
0.666
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr12_+_121416548
|
0.665
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr16_+_67282774
|
0.663
|
NM_004594
|
SLC9A5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5
|
|
chr3_-_50378238
|
0.661
|
NM_001206957
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr19_-_15560761
|
0.660
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|