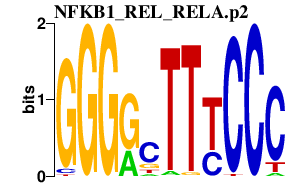
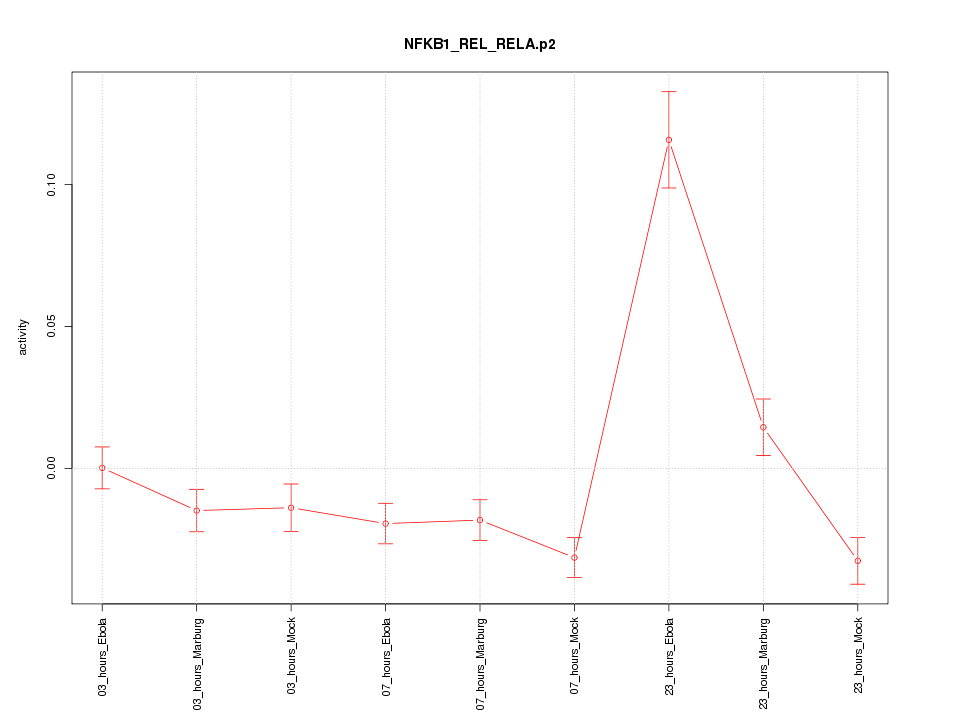
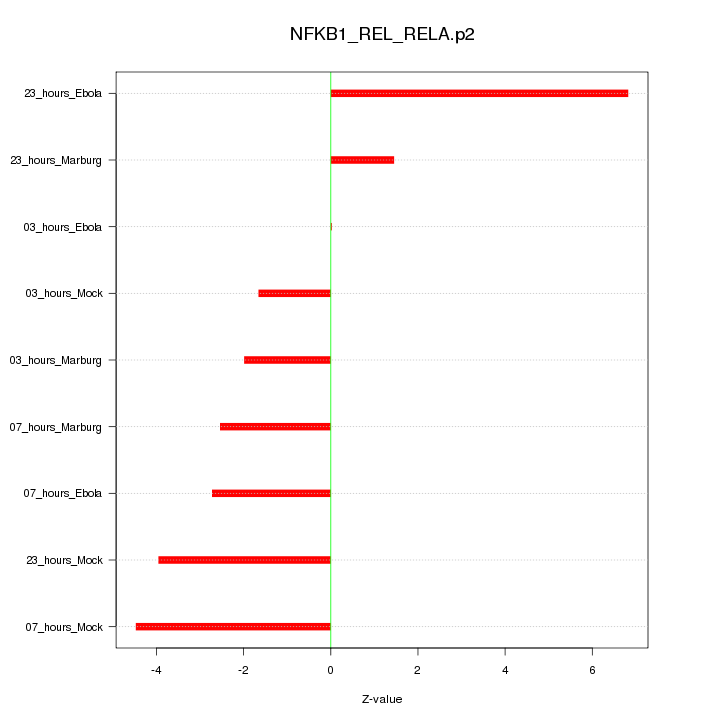
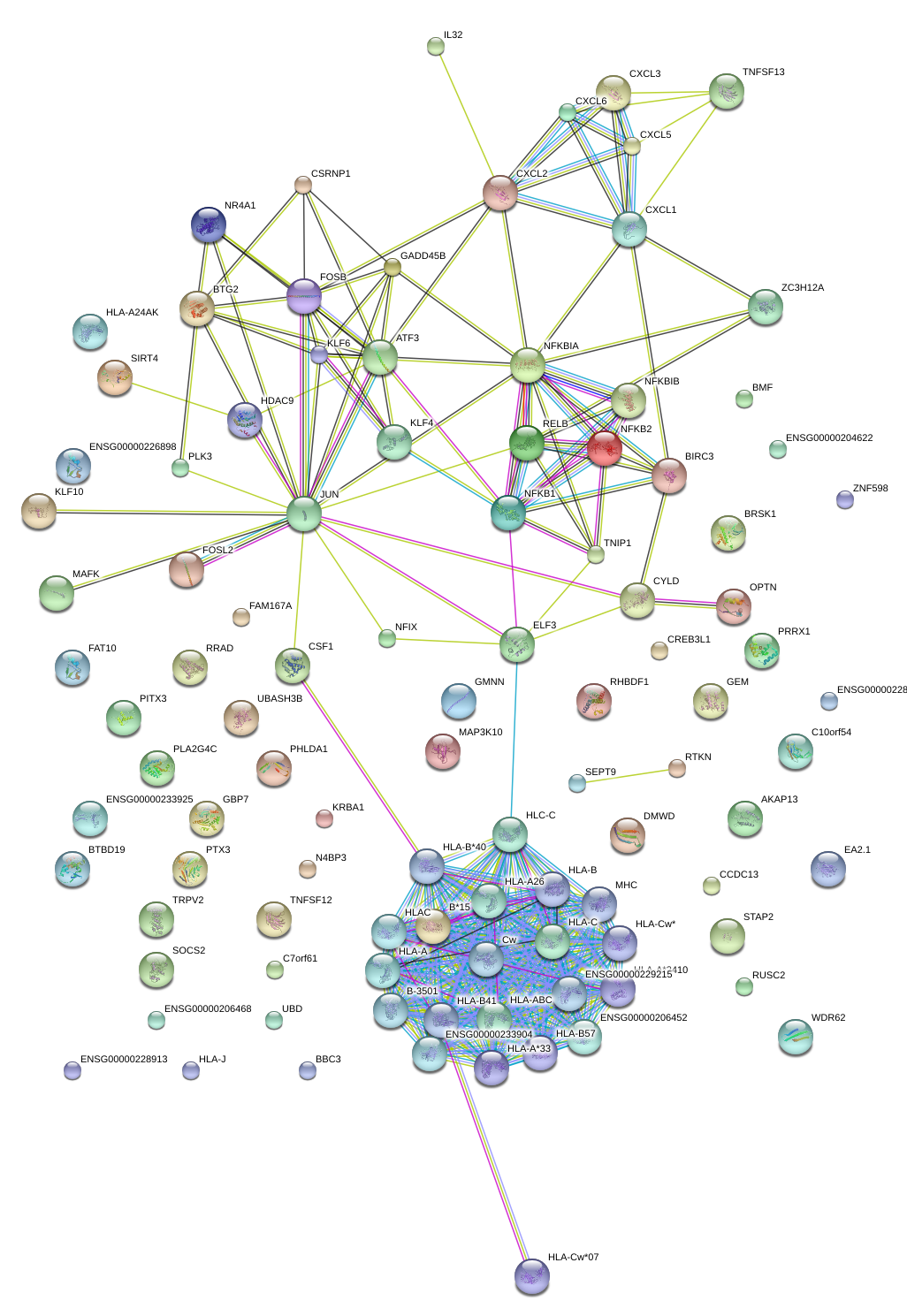
|
chr10_+_104154228
|
9.600
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr10_+_104155431
|
7.542
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr4_-_74964836
|
6.720
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr7_+_18548899
|
6.353
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr19_+_45504686
|
5.747
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr16_-_66959418
|
5.271
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr9_+_35560424
|
5.151
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_18548963
|
5.018
|
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_1570321
|
4.972
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr8_-_103667882
|
4.749
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr4_-_74864296
|
4.728
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr4_+_74735107
|
4.481
|
NM_001511
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr12_+_52445185
|
4.457
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr8_-_95274540
|
4.320
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr4_-_74964996
|
4.303
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr19_+_45971249
|
4.302
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr12_-_76425375
|
4.279
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr11_+_122526339
|
4.111
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr12_-_76425207
|
4.032
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr11_+_102188180
|
4.000
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr4_+_74702272
|
3.835
|
NM_002993
|
CXCL6
|
chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2)
|
|
chr8_-_95274520
|
3.781
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr19_-_47734215
|
3.711
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr19_+_2476122
|
3.597
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr19_-_4338750
|
3.596
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr19_+_40697516
|
3.560
|
NM_002446
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10
|
|
chr4_-_74904329
|
3.550
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr16_+_3115639
|
3.515
|
|
IL32
|
interleukin 32
|
|
chr1_+_201979689
|
3.505
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr1_+_201979747
|
3.385
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr1_+_45274153
|
3.371
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr16_+_3115312
|
3.348
|
NM_001012631
NM_001012633
NM_001012718
NM_004221
|
IL32
|
interleukin 32
|
|
chr10_-_73533196
|
3.284
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr1_+_35331533
|
3.280
|
|
|
|
|
chr17_+_7461591
|
3.254
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr10_-_3827205
|
3.240
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr1_+_170633312
|
3.207
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr19_-_48614013
|
3.180
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr1_+_110453232
|
3.151
|
NM_000757
NM_172210
NM_172212
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr16_+_50775960
|
3.137
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr15_+_85923739
|
3.131
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr1_+_110453456
|
3.126
|
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr7_-_100061893
|
3.042
|
NM_001004323
|
C7orf61
|
chromosome 7 open reading frame 61
|
|
chr12_+_120740123
|
2.988
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr2_+_28615668
|
2.988
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr1_-_59248287
|
2.943
|
|
JUN
|
jun proto-oncogene
|
|
chr1_+_45266035
|
2.929
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr6_+_29974352
|
2.913
|
|
HLA-J
|
major histocompatibility complex, class I, J (pseudogene)
|
|
chr1_-_1475739
|
2.892
|
NM_001114748
|
TMEM240
|
transmembrane protein 240
|
|
chr16_-_2059759
|
2.857
|
NM_178167
|
ZNF598
|
zinc finger protein 598
|
|
chr3_+_157154579
|
2.854
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr3_-_42814734
|
2.849
|
NM_144719
|
CCDC13
|
coiled-coil domain containing 13
|
|
chr1_+_45266048
|
2.821
|
|
PLK3
|
polo-like kinase 3
|
|
chr2_-_74669005
|
2.820
|
|
RTKN
|
rhotekin
|
|
chr19_+_39390618
|
2.819
|
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr8_-_11324184
|
2.817
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr10_+_13142055
|
2.816
|
NM_001008211
NM_001008212
NM_001008213
NM_021980
|
OPTN
|
optineurin
|
|
chr4_+_103422730
|
2.805
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr16_+_3115611
|
2.802
|
NM_001012635
|
IL32
|
interleukin 32
|
|
chr6_-_29527701
|
2.759
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr1_+_37940180
|
2.742
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr19_+_39390570
|
2.701
|
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr16_+_3115638
|
2.691
|
NM_001012634
|
IL32
|
interleukin 32
|
|
chr19_+_13205956
|
2.620
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr16_-_122573
|
2.610
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr6_-_29527477
|
2.601
|
|
UBD
|
ubiquitin D
|
|
chr5_+_177540465
|
2.581
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr4_+_103422822
|
2.579
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr16_+_50776012
|
2.558
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr17_+_16318848
|
2.551
|
NM_016113
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr11_+_46299554
|
2.514
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr9_-_110250707
|
2.512
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr19_+_39390258
|
2.499
|
NM_001243116
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr7_+_149412027
|
2.464
|
NM_032534
|
KRBA1
|
KRAB-A domain containing 1
|
|
chr3_-_39195101
|
2.450
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr1_-_89641722
|
2.432
|
NM_207398
|
GBP7
|
guanylate binding protein 7
|
|
chr1_+_203274646
|
2.429
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr4_+_103422479
|
2.403
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr15_-_40398286
|
2.391
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr19_-_46295647
|
2.385
|
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr14_-_35873823
|
2.359
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr2_-_74669026
|
2.356
|
|
RTKN
|
rhotekin
|
|
chr19_+_55795533
|
2.353
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr1_+_212788359
|
2.351
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr19_-_46283281
|
2.334
|
|
|
|
|
chr6_-_31324955
|
2.333
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr4_+_103422553
|
2.309
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr19_+_55795766
|
2.287
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr12_+_93964801
|
2.284
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr5_-_150466696
|
2.253
|
NM_001252391
NM_001252392
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr3_-_39195034
|
2.245
|
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr22_-_50708720
|
2.215
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr17_+_16318911
|
2.188
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr2_+_228678556
|
2.161
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr17_-_39092714
|
2.149
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr19_-_4831711
|
2.130
|
NM_182919
|
TICAM1
|
toll-like receptor adaptor molecule 1
|
|
chr18_+_23806385
|
2.118
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr20_-_44600787
|
2.083
|
NM_022095
|
ZNF335
|
zinc finger protein 335
|
|
chr5_+_176875325
|
2.079
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr20_-_44600939
|
2.039
|
|
ZNF335
|
zinc finger protein 335
|
|
chr3_-_156878481
|
2.024
|
NM_020307
|
CCNL1
|
cyclin L1
|
|
chr19_+_827821
|
2.011
|
NM_001700
|
AZU1
|
azurocidin 1
|
|
chr7_+_17338182
|
2.005
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr22_-_38506532
|
1.997
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr7_+_150745604
|
1.996
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr11_+_46299161
|
1.973
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr22_+_38599026
|
1.950
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr5_+_60628085
|
1.936
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr11_+_59522905
|
1.917
|
|
STX3
|
syntaxin 3
|
|
chr10_-_125851850
|
1.914
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr11_+_65984213
|
1.897
|
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr6_+_17281773
|
1.890
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr12_-_58160823
|
1.889
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr19_+_48673936
|
1.875
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr2_-_74669055
|
1.874
|
NM_001015055
|
RTKN
|
rhotekin
|
|
chr2_+_61108702
|
1.865
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr6_+_138188352
|
1.859
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr6_+_29910246
|
1.849
|
NM_001242758
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr14_+_75988765
|
1.845
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr8_+_128806720
|
1.827
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr6_+_108882068
|
1.827
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr7_-_100808832
|
1.823
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr16_+_67571342
|
1.821
|
NM_001193524
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr11_+_59522801
|
1.818
|
|
STX3
|
syntaxin 3
|
|
chr6_+_31540092
|
1.815
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr2_+_61108724
|
1.814
|
NM_002908
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr17_+_38474513
|
1.813
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr2_+_43864405
|
1.809
|
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr19_-_40724272
|
1.803
|
NM_152479
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr6_-_150039219
|
1.798
|
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr6_+_108882104
|
1.793
|
|
FOXO3
|
forkhead box O3
|
|
chr17_+_38474472
|
1.785
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr9_-_86432546
|
1.779
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr6_+_36646455
|
1.772
|
NM_000389
NM_001220778
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr11_+_6625000
|
1.768
|
|
ILK
|
integrin-linked kinase
|
|
chr20_-_3388586
|
1.757
|
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr17_+_38474544
|
1.750
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr1_-_202129731
|
1.743
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr13_+_42614171
|
1.742
|
NM_001204504
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr12_-_15942116
|
1.725
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr5_+_172571444
|
1.721
|
NM_001205
NM_013978
NM_013979
NM_013980
|
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1
|
|
chr19_-_1513187
|
1.712
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr22_-_18484183
|
1.705
|
|
MICAL3
|
microtubule associated monoxygenase, calponin and LIM domain containing 3
|
|
chr19_-_41859830
|
1.693
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr9_+_130860769
|
1.690
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr19_+_48111437
|
1.686
|
NM_015711
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1
|
|
chr5_-_150460550
|
1.683
|
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr7_-_104654049
|
1.681
|
|
LOC100216545
|
uncharacterized LOC100216545
|
|
chr1_+_162039469
|
1.679
|
NM_001164757
NM_014697
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr13_+_21872248
|
1.678
|
|
MIPEPP3
|
mitochondrial intermediate peptidase pseudogene 3
|
|
chr12_-_15942350
|
1.675
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr11_+_46299585
|
1.675
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr2_+_61108775
|
1.661
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr15_+_76352270
|
1.658
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr5_-_148758787
|
1.644
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr5_-_150460599
|
1.644
|
NM_001252385
NM_001252386
NM_001252393
NM_006058
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr10_-_116527955
|
1.644
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr14_-_65879245
|
1.635
|
|
LOC645431
|
uncharacterized LOC645431
|
|
chr20_-_3388219
|
1.630
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr11_+_6625206
|
1.629
|
NM_001014795
|
ILK
|
integrin-linked kinase
|
|
chr11_+_66059364
|
1.625
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr19_+_56152281
|
1.614
|
NM_207115
|
ZNF580
|
zinc finger protein 580
|
|
chr13_+_51796363
|
1.613
|
NM_001242312
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr19_+_41222915
|
1.613
|
NM_025194
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr5_+_612454
|
1.613
|
|
CEP72
|
centrosomal protein 72kDa
|
|
chr6_+_41514133
|
1.609
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr5_+_172484353
|
1.595
|
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr2_-_206950895
|
1.594
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr16_-_54318889
|
1.581
|
|
IRX3
|
iroquois homeobox 3
|
|
chr1_-_156675237
|
1.575
|
NM_001878
NM_001199723
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr1_-_236030170
|
1.572
|
|
|
|
|
chr10_-_32667543
|
1.568
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr9_-_110251308
|
1.568
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr19_+_39390567
|
1.565
|
NM_002503
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr6_+_41514289
|
1.560
|
|
FOXP4
|
forkhead box P4
|
|
chr14_+_24616095
|
1.557
|
|
RNF31
|
ring finger protein 31
|
|
chr19_+_13135809
|
1.552
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr2_-_211035913
|
1.551
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr2_+_43864433
|
1.542
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr10_-_98273444
|
1.540
|
NM_012465
|
TLL2
|
tolloid-like 2
|
|
chr4_+_139936938
|
1.539
|
NM_012118
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae)
|
|
chr17_+_45973515
|
1.537
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr11_+_66059335
|
1.531
|
|
TMEM151A
|
transmembrane protein 151A
|
|
chr19_+_1067536
|
1.530
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr17_-_18907988
|
1.526
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr6_+_36646489
|
1.525
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr5_+_612404
|
1.524
|
NM_018140
|
CEP72
|
centrosomal protein 72kDa
|
|
chr21_-_47705235
|
1.515
|
NM_003906
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr2_-_96700662
|
1.513
|
NM_207328
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial
|
|
chr3_-_50605062
|
1.506
|
NM_001171740
NM_001171743
NM_016210
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr19_+_52839497
|
1.500
|
NM_001161425
NM_001161426
NM_001161427
|
ZNF610
|
zinc finger protein 610
|
|
chr6_-_109775423
|
1.495
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr22_-_37640287
|
1.493
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr9_+_130860816
|
1.490
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr3_+_19189969
|
1.480
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr5_-_150467220
|
1.477
|
NM_001252390
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr4_-_82392971
|
1.474
|
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr19_+_13135745
|
1.473
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr6_-_150039391
|
1.470
|
NM_004690
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr13_-_31735718
|
1.466
|
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|