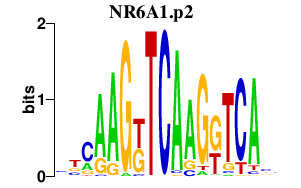
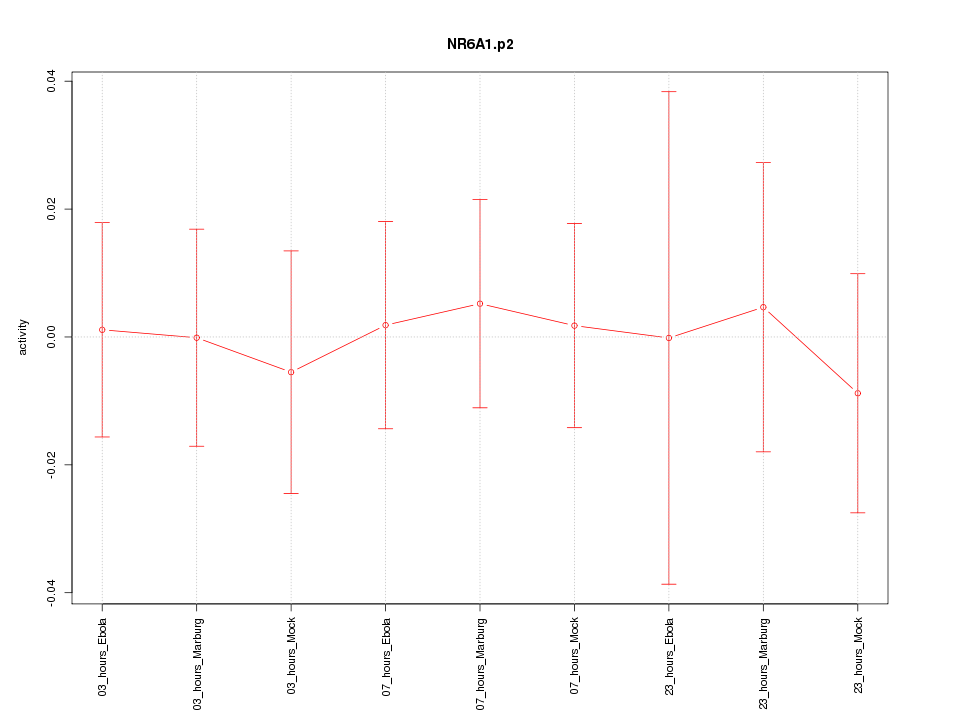
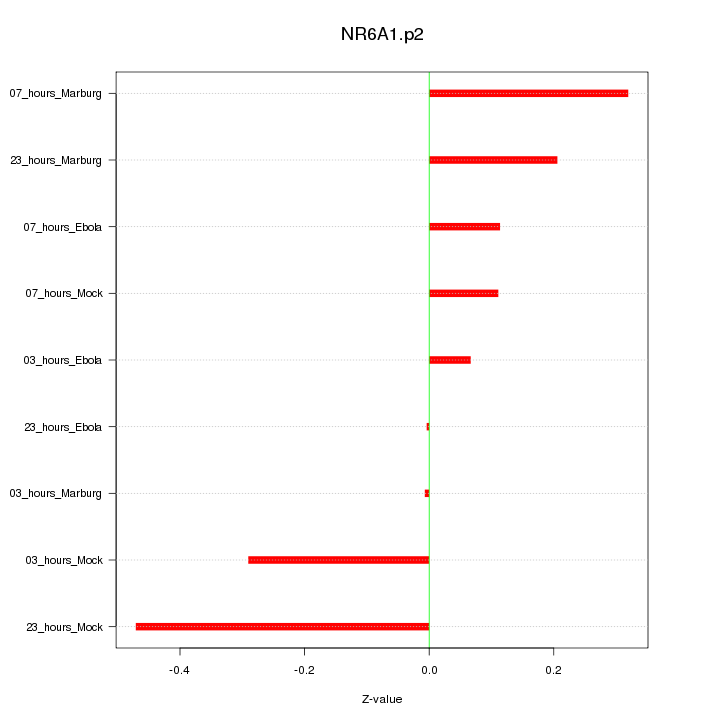
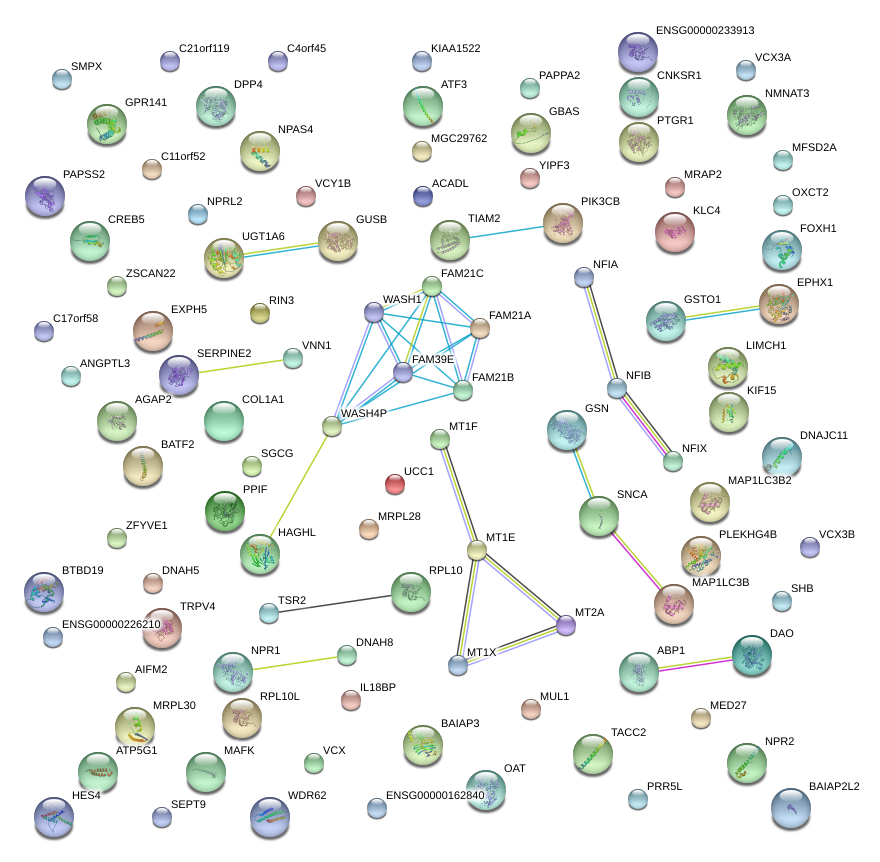
|
chr12_+_7169192
|
0.131
|
|
|
|
|
chr16_+_776957
|
0.118
|
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr7_+_28530747
|
0.113
|
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_+_111789658
|
0.100
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr11_+_111789600
|
0.091
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr9_-_38069082
|
0.080
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr6_-_133035180
|
0.072
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr2_-_224903324
|
0.064
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr1_-_935469
|
0.063
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr10_-_71892514
|
0.063
|
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2
|
|
chr2_-_162930629
|
0.062
|
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr1_+_176432306
|
0.061
|
NM_020318
NM_021936
|
PAPPA2
|
pappalysin 2
|
|
chr10_+_89419352
|
0.054
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr16_+_56642477
|
0.054
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr2_-_162930500
|
0.047
|
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr1_+_19923457
|
0.040
|
NM_001204088
NM_001204089
NM_001032363
NM_001204082
NM_001204083
|
C1orf151-NBL1
MINOS1
|
C1orf151-NBL1 readthrough
mitochondrial inner membrane organizing system 1
|
|
chr10_-_71892596
|
0.040
|
NM_001198696
NM_032797
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2
|
|
chr10_+_123754141
|
0.039
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr9_+_35792405
|
0.038
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr21_+_33765411
|
0.037
|
|
C21orf119
|
chromosome 21 open reading frame 119
|
|
chr2_-_162930544
|
0.033
|
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr11_+_65192284
|
0.033
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr13_+_23755059
|
0.033
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr11_-_64764388
|
0.033
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chr2_-_200715895
|
0.032
|
|
FONG
|
uncharacterized LOC348751
|
|
chr7_+_37960418
|
0.032
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr2_-_211090146
|
0.030
|
NM_001608
|
ACADL
|
acyl-CoA dehydrogenase, long chain
|
|
chr9_-_38069207
|
0.028
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr1_-_6761839
|
0.028
|
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr16_-_420508
|
0.027
|
NM_006428
|
MRPL28
|
mitochondrial ribosomal protein L28
|
|
chr10_+_106013951
|
0.027
|
NM_001191003
|
GSTO1
|
glutathione S-transferase omega 1
|
|
chr12_-_110252600
|
0.027
|
NM_001177431
NM_001177428
NM_001177433
NM_147204
|
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr11_+_66188474
|
0.026
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chrX_-_21776158
|
0.025
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr1_-_20834516
|
0.024
|
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chrX_+_8432870
|
0.024
|
NM_001001888
|
VCX3B
VCX3A
|
variable charge, X-linked 3B
variable charge, X-linked 3A
|
|
chr9_-_114362100
|
0.023
|
NM_001146108
NM_001146109
|
PTGR1
|
prostaglandin reductase 1
|
|
chr1_+_225997742
|
0.023
|
NM_001136018
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr12_+_116997185
|
0.023
|
NM_001085481
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2
|
|
chr6_+_155411422
|
0.022
|
NM_012454
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr3_-_139396773
|
0.020
|
NM_001200047
NM_178177
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr1_+_33231260
|
0.020
|
|
KIAA1522
|
KIAA1522
|
|
chr9_-_14313899
|
0.019
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_225997804
|
0.019
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr17_-_48264039
|
0.019
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr9_-_29738
|
0.018
|
NM_182905
|
WASH1
|
WAS protein family homolog 1
|
|
chr14_-_47120940
|
0.017
|
NM_080746
|
RPL10L
|
ribosomal protein L10-like
|
|
chr1_+_26503973
|
0.017
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr20_-_43024303
|
0.017
|
|
|
|
|
chr1_-_20834609
|
0.016
|
NM_024544
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chr9_-_14313943
|
0.016
|
|
NFIB
|
nuclear factor I/B
|
|
chr4_+_41614725
|
0.016
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_-_134955056
|
0.016
|
|
MED27
|
mediator complex subunit 27
|
|
chr1_-_40236926
|
0.015
|
NM_022120
|
OXCT2
|
3-oxoacid CoA transferase 2
|
|
chr22_-_24059436
|
0.015
|
|
|
|
|
chr7_+_1577118
|
0.014
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr11_-_108464344
|
0.013
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr12_-_58135853
|
0.012
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr3_-_138478184
|
0.012
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr2_-_162929928
|
0.011
|
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr5_-_14011846
|
0.011
|
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr17_+_46970128
|
0.011
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chrX_+_54466840
|
0.011
|
NM_058163
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr1_+_40420802
|
0.011
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr7_+_56032065
|
0.010
|
NM_001202469
NM_001483
|
GBAS
|
glioblastoma amplified sequence
|
|
chr22_-_24059524
|
0.010
|
|
GUSBP11
|
glucuronidase, beta pseudogene 11
|
|
chr7_-_65447206
|
0.010
|
|
GUSB
|
glucuronidase, beta
|
|
chr7_-_65447162
|
0.010
|
|
GUSB
|
glucuronidase, beta
|
|
chr4_-_90758126
|
0.009
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr8_-_145701139
|
0.009
|
|
FOXH1
|
forkhead box H1
|
|
chr1_+_63063186
|
0.009
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr6_+_43028181
|
0.009
|
NM_201522
|
KLC4
|
kinesin light chain 4
|
|
chr14_+_93118793
|
0.009
|
|
RIN3
|
Ras and Rab interactor 3
|
|
chr4_-_159956332
|
0.009
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr6_+_84743250
|
0.008
|
NM_138409
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr14_-_73453651
|
0.008
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr22_-_24059502
|
0.008
|
|
GUSBP11
|
glucuronidase, beta pseudogene 11
|
|
chr11_-_62472901
|
0.008
|
|
|
|
|
chr9_+_124043719
|
0.008
|
|
GSN
|
gelsolin
|
|
chr10_+_81107259
|
0.007
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr6_-_43484503
|
0.007
|
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr1_+_45274153
|
0.007
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr11_+_71709951
|
0.007
|
NM_001039659
NM_001039660
NM_173044
NM_005699
NM_173042
|
IL18BP
|
interleukin 18 binding protein
|
|
chr7_-_65447233
|
0.007
|
NM_000181
|
GUSB
|
glucuronidase, beta
|
|
chr12_+_109273856
|
0.006
|
NM_001917
|
DAO
|
D-amino-acid oxidase
|
|
chr11_+_36317724
|
0.006
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr16_+_1386197
|
0.006
|
NM_001199096
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr1_+_212738675
|
0.006
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr10_-_126107493
|
0.006
|
NM_000274
NM_001171814
|
OAT
|
ornithine aminotransferase
|
|
chr10_+_51837885
|
0.006
|
NM_018232
|
FAM21B
|
family with sequence similarity 21, member B
|
|
chrX_+_17653412
|
0.005
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr1_-_161102457
|
0.005
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr1_+_40420837
|
0.005
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr3_+_46411632
|
0.005
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr18_+_33767420
|
0.005
|
NM_017947
|
MOCOS
|
molybdenum cofactor sulfurase
|
|
chr1_-_16134192
|
0.005
|
NM_001089591
|
UQCRHL
|
ubiquinol-cytochrome c reductase hinge protein-like
|
|
chr10_+_106014467
|
0.005
|
NM_001191002
NM_004832
|
GSTO1
|
glutathione S-transferase omega 1
|
|
chr5_-_41213513
|
0.005
|
NM_000065
|
C6
|
complement component 6
|
|
chr14_-_58893867
|
0.004
|
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast)
|
|
chr12_+_54410893
|
0.004
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr6_+_117803779
|
0.004
|
NM_173674
|
DCBLD1
|
discoidin, CUB and LCCL domain containing 1
|
|
chr7_-_2883649
|
0.004
|
NM_007353
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12
|
|
chr6_-_56407681
|
0.004
|
|
|
|
|
chr5_+_140749830
|
0.004
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr1_+_117452359
|
0.004
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr20_-_3149070
|
0.003
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr19_-_5846465
|
0.003
|
|
FUT3
|
fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group)
|
|
chr15_-_41166336
|
0.003
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr11_-_796175
|
0.003
|
NM_024698
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr8_-_135725050
|
0.003
|
|
|
|
|
chr5_-_14871555
|
0.003
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr10_-_126107454
|
0.003
|
|
OAT
|
ornithine aminotransferase
|
|
chr6_-_43484701
|
0.003
|
NM_015388
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr3_-_38178463
|
0.003
|
|
ACAA1
|
acetyl-CoA acyltransferase 1
|
|
chr1_-_120612260
|
0.003
|
|
NOTCH2
|
notch 2
|
|
chr8_-_145701717
|
0.003
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr2_+_33172368
|
0.003
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chrX_+_48916515
|
0.003
|
NM_001163322
NM_033626
NM_001163321
NM_001163323
|
CCDC120
|
coiled-coil domain containing 120
|
|
chr1_+_113236679
|
0.003
|
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse)
|
|
chr2_+_191002485
|
0.002
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr1_-_120612175
|
0.002
|
|
NOTCH2
|
notch 2
|
|
chr7_+_37960162
|
0.002
|
NM_001242946
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr3_-_14989857
|
0.002
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr1_-_160617042
|
0.002
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr16_+_3451154
|
0.002
|
NM_001032292
NM_003450
|
ZNF174
|
zinc finger protein 174
|
|
chr1_+_6684924
|
0.001
|
NM_001195753
|
THAP3
|
THAP domain containing, apoptosis associated protein 3
|
|
chr17_+_12692779
|
0.001
|
NM_014859
|
ARHGAP44
|
Rho GTPase activating protein 44
|
|
chr22_+_51041561
|
0.001
|
NM_016431
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr7_-_87936202
|
0.001
|
NM_001205315
NM_001205316
NM_024636
|
STEAP4
|
STEAP family member 4
|
|
chr20_+_17680592
|
0.001
|
NM_001014977
NM_178477
|
BANF2
|
barrier to autointegration factor 2
|
|
chr5_-_60458228
|
0.001
|
NM_001048249
|
C5orf43
|
chromosome 5 open reading frame 43
|
|
chr12_-_6484389
|
0.001
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr9_+_139981338
|
0.001
|
|
MAN1B1
|
mannosidase, alpha, class 1B, member 1
|
|
chr7_-_126883568
|
0.001
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr4_-_89205667
|
0.001
|
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K
|
|
chr7_-_100240334
|
0.000
|
|
TFR2
|
transferrin receptor 2
|
|
chr16_-_28222700
|
0.000
|
|
XPO6
|
exportin 6
|