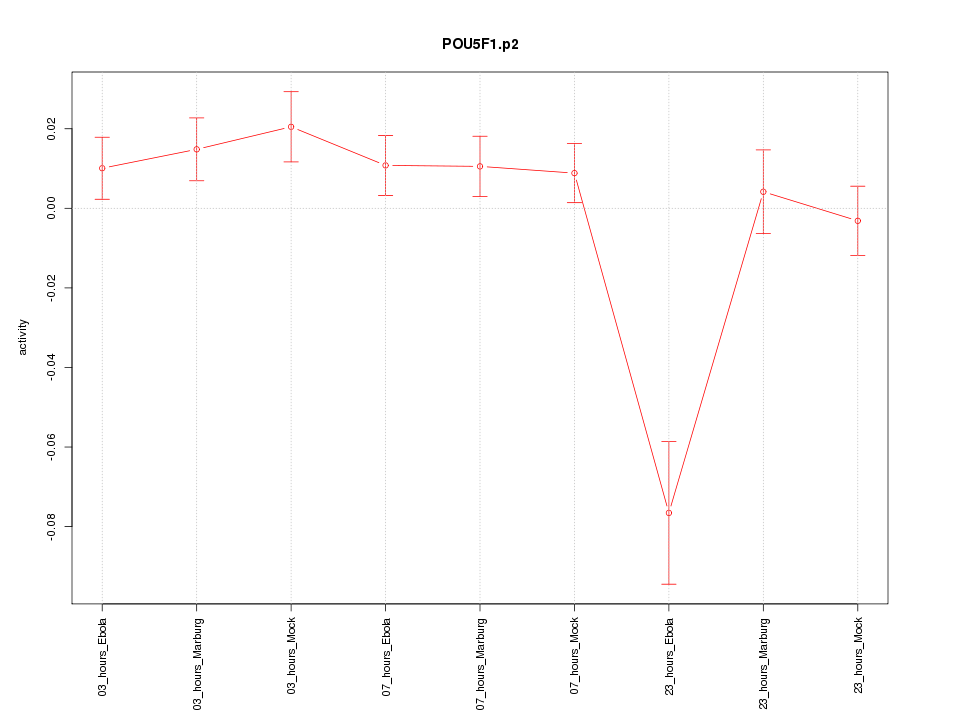
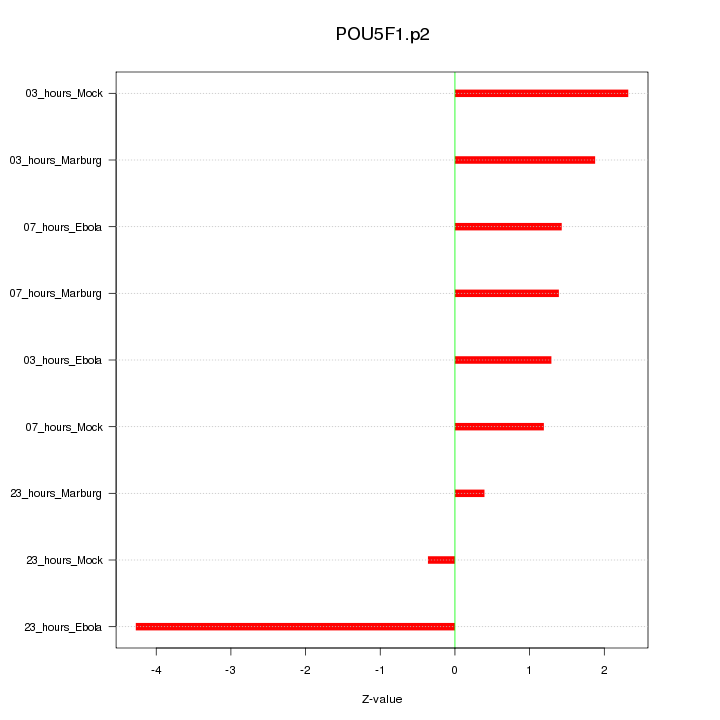
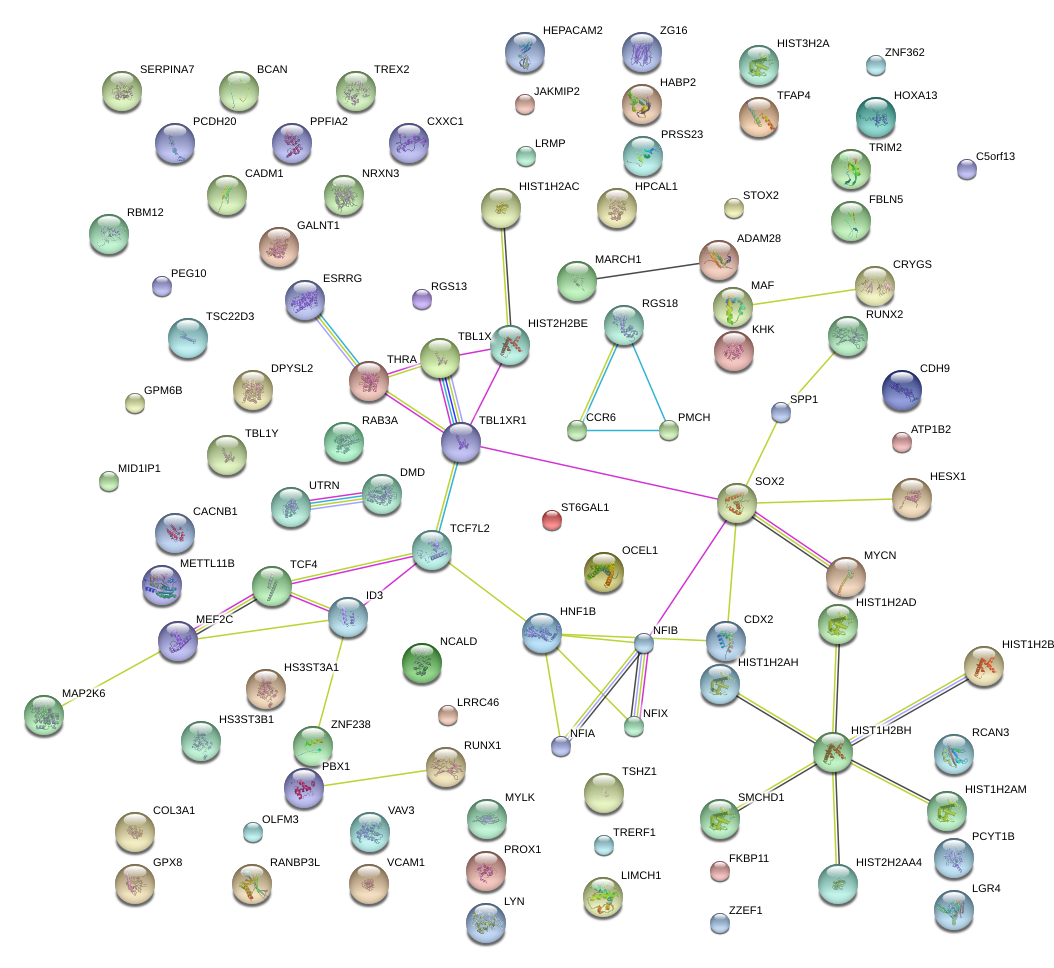
|
chr17_+_67410825
|
18.375
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr10_+_115312777
|
6.277
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr1_+_164528914
|
5.123
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr5_-_36301999
|
4.249
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr17_-_36104874
|
3.996
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr17_+_7554415
|
3.821
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr6_+_27100784
|
3.690
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr5_-_111091947
|
3.565
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr14_+_79745653
|
3.565
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr21_-_36421195
|
3.510
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_217262946
|
3.504
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_+_26124372
|
3.216
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr6_+_26251826
|
3.194
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr5_-_27038650
|
3.176
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr3_-_123603144
|
3.104
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr17_-_36105005
|
2.992
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr5_+_54455945
|
2.855
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr11_-_27494232
|
2.779
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr1_+_170115187
|
2.680
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr14_-_92413738
|
2.595
|
|
FBLN5
|
fibulin 5
|
|
chr20_-_34243118
|
2.592
|
|
RBM12
|
RNA binding motif protein 12
|
|
chrX_+_38660692
|
2.566
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chrX_+_38660653
|
2.513
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chrX_-_33357725
|
2.486
|
NM_000109
|
DMD
|
dystrophin
|
|
chr2_+_16080559
|
2.470
|
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr16_-_79634610
|
2.456
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr1_+_33722158
|
2.448
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chrX_-_24665225
|
2.352
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr8_-_102803196
|
2.339
|
|
NCALD
|
neurocalcin delta
|
|
chr5_-_147162056
|
2.328
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr19_-_18314731
|
2.303
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr1_+_192605267
|
2.257
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr1_-_108231122
|
2.206
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr8_+_26435590
|
2.205
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr2_+_16080639
|
2.203
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr3_+_181429711
|
2.185
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chrX_+_9431314
|
2.175
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr17_+_7554239
|
2.136
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chrX_-_24665454
|
2.114
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr11_-_115375065
|
2.088
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr16_+_29789560
|
2.070
|
NM_152338
|
ZG16
|
zymogen granule protein 16 homolog (rat)
|
|
chr13_-_28543143
|
2.061
|
|
CDX2
|
caudal type homeobox 2
|
|
chr1_+_24840800
|
2.049
|
NM_001251980
NM_001251982
NM_001251985
|
RCAN3
|
RCAN family member 3
|
|
chr1_+_39875175
|
2.022
|
NM_015038
|
KIAA0754
|
KIAA0754
|
|
chr21_-_36421461
|
2.016
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr16_-_4323000
|
2.004
|
NM_003223
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr17_+_38219040
|
1.995
|
NM_001190918
NM_003250
NM_199334
|
THRA
|
thyroid hormone receptor, alpha
|
|
chr6_+_167536240
|
1.960
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr13_-_61989209
|
1.957
|
|
PCDH20
|
protocadherin 20
|
|
chrX_-_105282714
|
1.943
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr1_+_164528586
|
1.922
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr2_+_27310079
|
1.909
|
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr17_+_7554711
|
1.897
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr5_-_111092918
|
1.864
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_-_21807120
|
1.853
|
|
|
|
|
chr6_-_42419782
|
1.846
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr4_+_41614911
|
1.840
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_111092865
|
1.800
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_-_13835313
|
1.785
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr12_+_25205488
|
1.780
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_-_81763127
|
1.777
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr1_-_23885916
|
1.770
|
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr18_-_53089722
|
1.761
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chrX_-_106960268
|
1.736
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr7_-_92855780
|
1.726
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr7_-_27239724
|
1.725
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr9_-_14086901
|
1.703
|
|
NFIB
|
nuclear factor I/B
|
|
chr3_-_186262114
|
1.700
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|
|
chr6_+_84222252
|
1.695
|
|
PRSS35
|
protease, serine, 35
|
|
chr17_+_45908992
|
1.687
|
NM_033413
|
LRRC46
|
leucine rich repeat containing 46
|
|
chr1_+_101185195
|
1.663
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr8_+_26435339
|
1.654
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr4_+_154178525
|
1.612
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr18_+_72922725
|
1.593
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr8_+_26435466
|
1.590
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr2_+_189839132
|
1.589
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr16_-_4322695
|
1.589
|
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr6_+_27114860
|
1.576
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr18_+_33234532
|
1.559
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr12_+_25205180
|
1.558
|
NM_001204126
NM_001204127
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr8_+_56792399
|
1.551
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr1_+_244214552
|
1.548
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr12_-_102591547
|
1.522
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr1_+_244214576
|
1.492
|
|
ZNF238
|
zinc finger protein 238
|
|
chr7_+_94292737
|
1.490
|
|
PEG10
|
paternally expressed 10
|
|
chr2_+_65663812
|
1.490
|
|
FLJ16124
|
FLJ16124 protein
|
|
chr1_-_102462371
|
1.476
|
|
OLFM3
|
olfactomedin 3
|
|
chr4_+_88896868
|
1.450
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_184826427
|
1.446
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr17_+_14204326
|
1.443
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr1_+_214161279
|
1.439
|
|
PROX1
|
prospero homeobox 1
|
|
chr4_+_88896838
|
1.433
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr5_-_88119604
|
1.432
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_164534656
|
1.428
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr5_-_111312522
|
1.425
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_186648273
|
1.411
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr8_+_24151579
|
1.397
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr3_-_57234279
|
1.396
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr19_+_17337012
|
1.391
|
NM_024578
|
OCEL1
|
occludin/ELL domain containing 1
|
|
chr11_+_63057371
|
1.376
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr6_-_26033795
|
1.347
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr3_+_49568370
|
1.339
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr2_-_188419185
|
1.338
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr13_-_28543423
|
1.337
|
|
CDX2
|
caudal type homeobox 2
|
|
chr6_+_112375277
|
1.335
|
NM_003880
NM_198239
|
WISP3
|
WNT1 inducible signaling pathway protein 3
|
|
chr11_-_94964209
|
1.335
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr9_+_132083294
|
1.334
|
NM_001012715
|
C9orf106
|
chromosome 9 open reading frame 106
|
|
chrX_-_13835213
|
1.324
|
|
GPM6B
|
glycoprotein M6B
|
|
chr6_-_52668599
|
1.315
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr4_-_186733372
|
1.310
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_-_21776158
|
1.306
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr4_+_41700736
|
1.303
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chrX_-_106960028
|
1.287
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_-_14989616
|
1.286
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr1_-_216896640
|
1.282
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_+_66458389
|
1.280
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr2_+_242127923
|
1.276
|
NM_001001666
NM_001001891
|
ANO7
|
anoctamin 7
|
|
chr10_-_42863435
|
1.271
|
|
LOC441666
|
zinc finger protein 91 pseudogene
|
|
chrX_+_22050920
|
1.253
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr1_+_87797301
|
1.244
|
|
|
|
|
chr2_-_152118258
|
1.244
|
NM_198557
|
RBM43
|
RNA binding motif protein 43
|
|
chr6_+_84222193
|
1.239
|
NM_001170423
NM_153362
|
PRSS35
|
protease, serine, 35
|
|
chr4_+_88896801
|
1.234
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_-_14989399
|
1.219
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chrM_-_379
|
1.218
|
|
|
|
|
chr16_+_30483959
|
1.217
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr6_-_134498973
|
1.209
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_-_115088628
|
1.205
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr4_-_141074122
|
1.205
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr3_-_71294229
|
1.201
|
NM_001244814
|
FOXP1
|
forkhead box P1
|
|
chr17_-_41276112
|
1.194
|
NM_007298
|
BRCA1
|
breast cancer 1, early onset
|
|
chr1_-_26185847
|
1.193
|
NM_024037
|
C1orf135
|
chromosome 1 open reading frame 135
|
|
chr5_-_147162251
|
1.192
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr8_+_56792345
|
1.189
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr1_-_211752052
|
1.188
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr2_-_165551178
|
1.186
|
|
COBLL1
|
COBL-like 1
|
|
chr14_+_90863302
|
1.176
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr16_+_2198607
|
1.171
|
NM_014353
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr11_+_5653484
|
1.166
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr13_-_28543290
|
1.166
|
NM_001265
|
CDX2
|
caudal type homeobox 2
|
|
chr5_-_73936299
|
1.157
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr16_+_30484049
|
1.153
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr7_+_43152196
|
1.153
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr7_+_141478241
|
1.152
|
NM_016944
|
TAS2R4
|
taste receptor, type 2, member 4
|
|
chr5_-_35230548
|
1.147
|
|
PRLR
|
prolactin receptor
|
|
chr1_+_92545861
|
1.137
|
NM_183242
|
BTBD8
|
BTB (POZ) domain containing 8
|
|
chr1_-_217250230
|
1.128
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_+_140602914
|
1.121
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr6_+_21593963
|
1.110
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr5_+_140743755
|
1.097
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr12_+_54447660
|
1.094
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr20_+_19915716
|
1.090
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr14_-_92414004
|
1.089
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr11_-_119599253
|
1.087
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr14_+_78870092
|
1.086
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr14_+_62462540
|
1.083
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr1_-_72748491
|
1.076
|
|
|
|
|
chr17_+_41052813
|
1.071
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr1_-_23886284
|
1.070
|
NM_002167
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr22_-_31742329
|
1.063
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr5_+_140593508
|
1.057
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr12_+_51318743
|
1.051
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr12_-_16760935
|
1.045
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_-_44123732
|
1.045
|
|
ZNF428
|
zinc finger protein 428
|
|
chr11_-_118084073
|
1.039
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr19_-_40562107
|
1.038
|
NM_001005851
|
ZNF780B
|
zinc finger protein 780B
|
|
chr7_-_150754934
|
1.033
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr12_-_81991959
|
1.031
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr3_+_3841079
|
1.031
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr21_-_36421630
|
1.028
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_+_88896899
|
1.025
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr19_-_44124013
|
1.023
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr6_-_134497069
|
1.021
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_-_6772252
|
1.018
|
NM_001127582
NM_001127583
NM_001127584
NM_001127585
NM_001127586
NM_016162
|
ING4
|
inhibitor of growth family, member 4
|
|
chr7_-_56101830
|
1.015
|
|
PSPH
|
phosphoserine phosphatase
|
|
chr20_+_34556528
|
1.015
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr2_+_232573226
|
1.009
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr2_+_232573249
|
1.008
|
|
PTMA
|
prothymosin, alpha
|
|
chr14_+_90863365
|
1.003
|
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr21_-_36421586
|
1.000
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_+_113739203
|
0.999
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr21_-_36421578
|
0.999
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_-_15402469
|
0.997
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr5_-_95158465
|
0.995
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr3_-_14989857
|
0.995
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr10_+_91061705
|
0.991
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chrX_-_63425484
|
0.986
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr2_+_234601511
|
0.985
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr1_+_81771844
|
0.985
|
|
LPHN2
|
latrophilin 2
|
|
chr2_-_188419049
|
0.982
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr4_-_139163222
|
0.978
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr15_+_96869156
|
0.977
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_+_172950207
|
0.971
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr19_-_46088074
|
0.970
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr12_-_63546589
|
0.967
|
NM_000706
|
AVPR1A
|
arginine vasopressin receptor 1A
|
|
chr11_-_66115016
|
0.967
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr6_-_26124130
|
0.966
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr4_-_141075232
|
0.958
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr10_-_91011586
|
0.956
|
NM_000235
NM_001127605
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr12_-_123920967
|
0.953
|
|
RILPL2
|
Rab interacting lysosomal protein-like 2
|