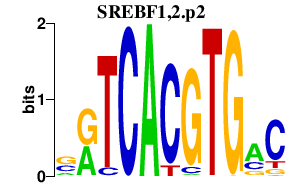
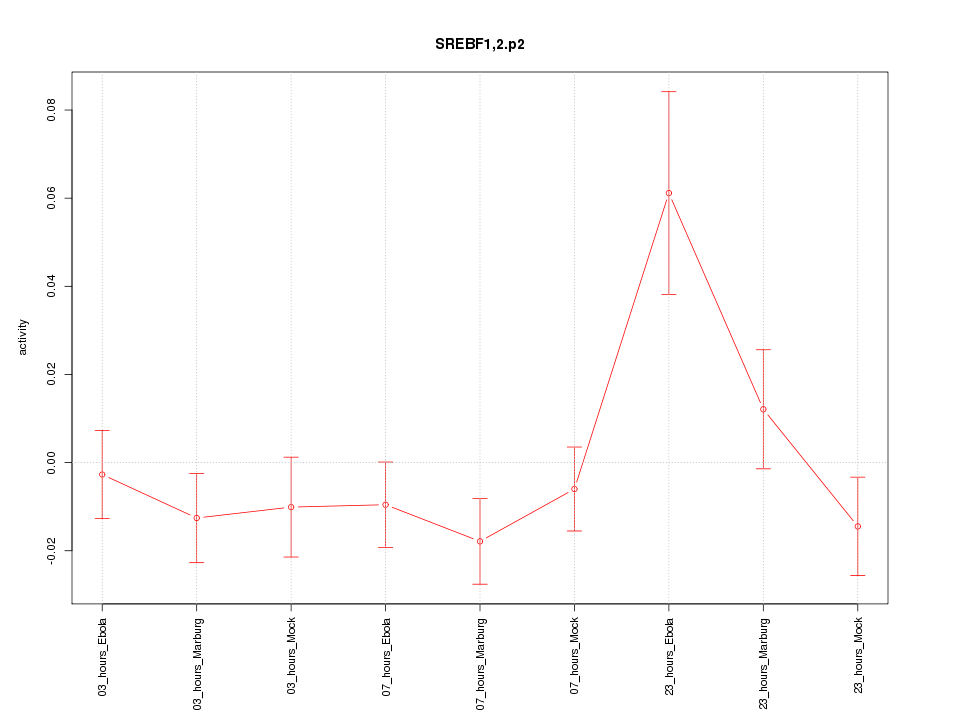
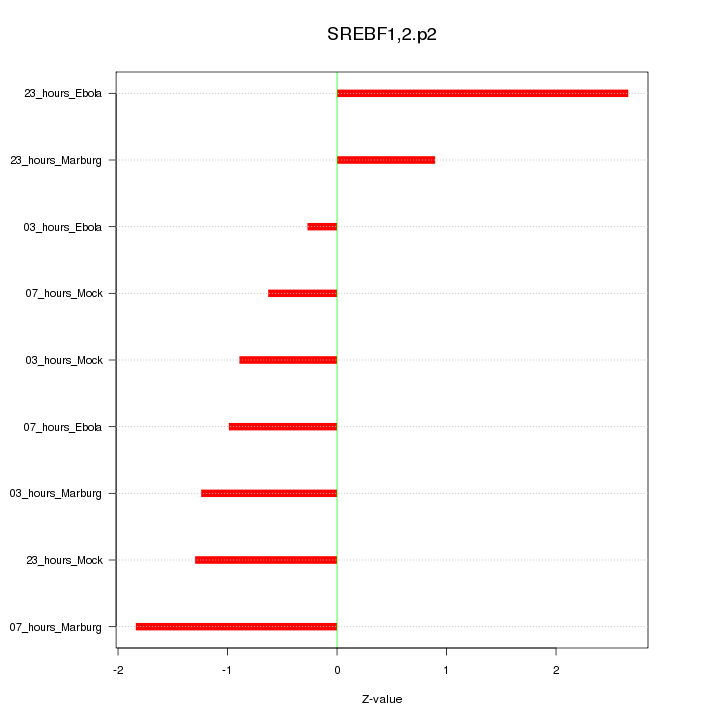
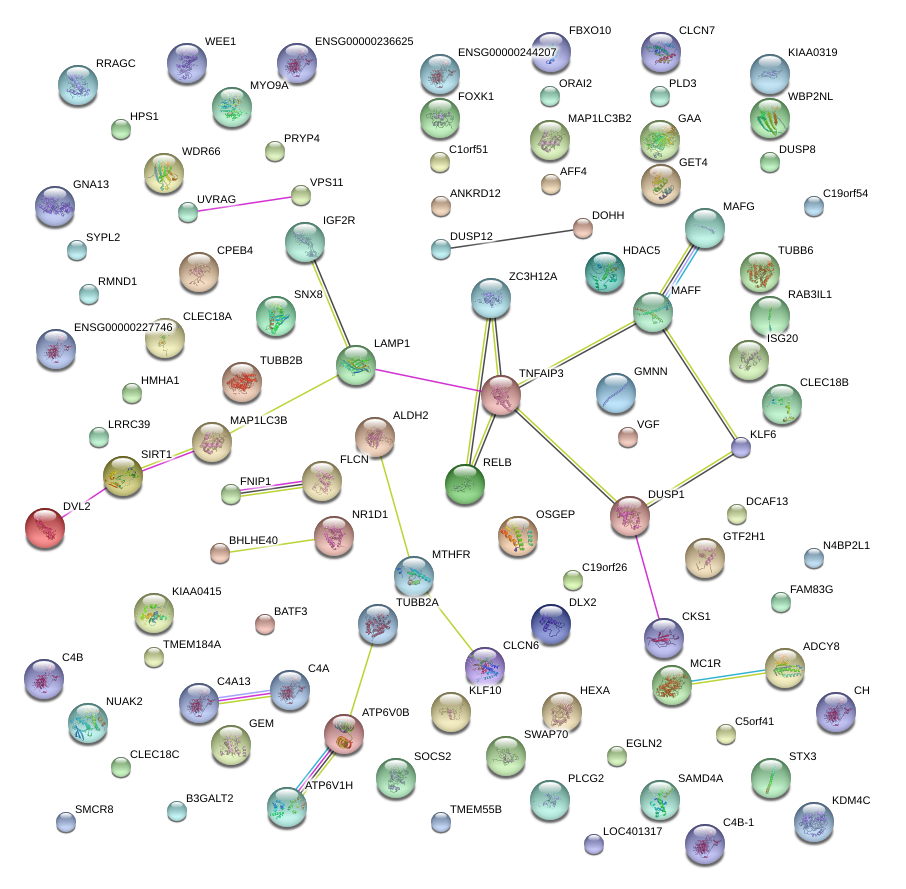
|
chr2_-_172967627
|
6.307
|
|
DLX2
|
distal-less homeobox 2
|
|
chr2_-_172967234
|
4.557
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr1_+_150255228
|
4.305
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr22_+_38599026
|
4.108
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr5_-_172198160
|
3.791
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr1_-_11866079
|
3.435
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr16_-_1525040
|
3.425
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr5_+_172483285
|
3.163
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr1_+_11866302
|
3.124
|
|
CLCN6
|
chloride channel 6
|
|
chr16_-_1525006
|
2.999
|
|
CLCN7
|
chloride channel 7
|
|
chr7_+_916178
|
2.830
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr7_+_28448970
|
2.747
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr16_-_1524926
|
2.697
|
|
CLCN7
|
chloride channel 7
|
|
chr8_-_95274540
|
2.571
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr1_-_39325317
|
2.524
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr16_-_1525020
|
2.454
|
|
CLCN7
|
chloride channel 7
|
|
chr19_+_45504686
|
2.386
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr7_+_4815258
|
2.322
|
NM_014855
|
KIAA0415
|
KIAA0415
|
|
chr8_-_95274520
|
2.292
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr10_-_3827418
|
2.292
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr1_+_11866206
|
2.285
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chrX_-_114797017
|
2.279
|
|
|
|
|
chr1_+_11866240
|
2.278
|
|
CLCN6
|
chloride channel 6
|
|
chr17_-_17140492
|
2.268
|
NM_144606
NM_144997
|
FLCN
|
folliculin
|
|
chr18_+_9136742
|
2.255
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr7_+_4815277
|
2.244
|
|
KIAA0415
|
KIAA0415
|
|
chr1_+_110009099
|
2.211
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr7_+_916240
|
2.131
|
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr5_-_131132690
|
2.100
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr12_+_93964801
|
2.087
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr11_+_9685650
|
2.062
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr10_-_3827205
|
2.055
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr11_+_9685624
|
2.036
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr5_-_131132715
|
2.017
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr5_-_131132642
|
2.001
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr17_-_7137675
|
1.941
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr9_-_37576248
|
1.900
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr11_+_9685606
|
1.873
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr19_+_1067293
|
1.869
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr19_-_1237840
|
1.855
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr16_+_89985268
|
1.838
|
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor)
|
|
chr7_-_100808832
|
1.824
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr8_-_54755836
|
1.821
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr5_-_131132709
|
1.788
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr10_+_69644341
|
1.784
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr11_+_9685653
|
1.767
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr11_+_18343794
|
1.749
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr6_+_31982538
|
1.736
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr11_+_9595451
|
1.714
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr17_-_79885544
|
1.704
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr17_-_18907988
|
1.704
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr3_+_5021096
|
1.700
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr17_-_7137580
|
1.661
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr8_-_54755821
|
1.660
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr11_+_9595429
|
1.650
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr19_-_41256371
|
1.650
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr1_-_11865963
|
1.635
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr6_+_138188352
|
1.634
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr12_+_122356446
|
1.634
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr2_-_220042731
|
1.609
|
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr11_+_59522801
|
1.591
|
|
STX3
|
syntaxin 3
|
|
chr11_-_1593149
|
1.587
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr10_-_100206657
|
1.586
|
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr15_+_89182038
|
1.582
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr11_-_61684981
|
1.578
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr8_-_54755546
|
1.571
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr17_-_42200957
|
1.543
|
|
HDAC5
|
histone deacetylase 5
|
|
chr3_+_5021141
|
1.541
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr17_-_42201009
|
1.535
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr11_+_75526211
|
1.510
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr1_-_205290756
|
1.497
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr1_+_44440642
|
1.494
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr10_+_69644708
|
1.485
|
|
SIRT1
|
sirtuin 1
|
|
chr11_+_9595309
|
1.476
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr19_-_3500620
|
1.466
|
NM_031304
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase
|
|
chr22_+_42394728
|
1.465
|
NM_152613
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr16_+_69984809
|
1.462
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr7_+_102074002
|
1.462
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr17_-_63052884
|
1.456
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr15_-_72668301
|
1.454
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr7_-_2354010
|
1.438
|
|
SNX8
|
sorting nexin 8
|
|
chr11_+_118938530
|
1.428
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr14_-_20929635
|
1.417
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr8_-_103667882
|
1.408
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr11_+_9595183
|
1.406
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr11_+_18344113
|
1.401
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr10_-_100206698
|
1.393
|
NM_000195
NM_182639
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr17_+_78075299
|
1.377
|
NM_000152
NM_001079803
NM_001079804
|
GAA
|
glucosidase, alpha; acid
|
|
chr17_-_7137722
|
1.374
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr5_-_132299248
|
1.362
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr11_+_118938528
|
1.356
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr17_+_18218569
|
1.349
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr17_-_17140425
|
1.338
|
|
FLCN
|
folliculin
|
|
chr19_+_40854331
|
1.336
|
NM_001031696
NM_012268
|
PLD3
|
phospholipase D family, member 3
|
|
chr14_-_20923266
|
1.336
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr17_-_38256905
|
1.333
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr11_+_118938473
|
1.331
|
NM_021729
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr11_+_75526236
|
1.328
|
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr5_-_132299263
|
1.327
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr1_-_212873197
|
1.313
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr16_+_81812925
|
1.310
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr14_+_55168831
|
1.308
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_-_193155723
|
1.293
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr17_-_63052744
|
1.293
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr1_+_44440673
|
1.289
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr6_-_151773168
|
1.280
|
NM_017909
|
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae)
|
|
chr7_+_27135672
|
1.276
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr13_+_113951461
|
1.270
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_-_33002218
|
1.262
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr7_-_1595746
|
1.258
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr11_+_118938515
|
1.256
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr15_-_72410226
|
1.253
|
NM_006901
|
MYO9A
|
myosin IXA
|
|
chr9_+_6757636
|
1.247
|
NM_001146694
NM_001146695
NM_015061
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr19_+_41305100
|
1.242
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr1_-_212873306
|
1.238
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr16_+_87425946
|
1.236
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr1_+_37940180
|
1.233
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr1_-_100643770
|
1.230
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr3_+_5021305
|
1.229
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr17_-_63052816
|
1.227
|
|
|
|
|
chr19_+_41305041
|
1.224
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_+_41305333
|
1.224
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr6_+_160390128
|
1.222
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr12_-_105501410
|
1.219
|
|
LOC414300
|
uncharacterized LOC414300
|
|
chr19_+_40854529
|
1.219
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr5_+_173315313
|
1.218
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr1_+_44440623
|
1.212
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_-_17109630
|
1.209
|
NM_178836
|
PLD6
|
phospholipase D family, member 6
|
|
chr17_+_57408907
|
1.206
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr15_-_72410108
|
1.202
|
|
MYO9A
|
myosin IXA
|
|
chr19_-_45909566
|
1.200
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr17_+_18218608
|
1.198
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr17_+_18218605
|
1.196
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr2_+_26395959
|
1.196
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr14_-_68283293
|
1.187
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr11_-_85780088
|
1.177
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_-_2354050
|
1.172
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr11_-_6640644
|
1.164
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr1_+_44440595
|
1.160
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_+_62502860
|
1.159
|
|
CEP95
|
centrosomal protein 95kDa
|
|
chr15_-_72668184
|
1.157
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr19_-_12833461
|
1.156
|
NM_001136196
|
TNPO2
|
transportin 2
|
|
chr14_-_35591439
|
1.148
|
NM_017917
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr14_-_20929484
|
1.143
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr11_+_59522905
|
1.134
|
|
STX3
|
syntaxin 3
|
|
chr6_-_91006460
|
1.130
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr16_+_87425874
|
1.127
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr9_-_124855559
|
1.123
|
|
TTLL11
|
tubulin tyrosine ligase-like family, member 11
|
|
chr7_-_30029284
|
1.117
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr7_-_100493481
|
1.115
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr17_-_17109601
|
1.115
|
|
PLD6
|
phospholipase D family, member 6
|
|
chr13_-_21635629
|
1.110
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr11_-_67141647
|
1.099
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr19_+_47759802
|
1.097
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr6_-_31830551
|
1.096
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr17_-_63052707
|
1.094
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr7_-_27135524
|
1.087
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr17_-_38256548
|
1.082
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr19_-_14606893
|
1.081
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr19_-_41256349
|
1.079
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr13_-_21635703
|
1.077
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr2_-_26101329
|
1.076
|
|
ASXL2
|
additional sex combs like 2 (Drosophila)
|
|
chr19_-_40324747
|
1.064
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr18_-_46987016
|
1.059
|
NM_017653
|
DYM
|
dymeclin
|
|
chr17_+_45401269
|
1.054
|
NM_001195192
NM_152347
|
C17orf57
|
chromosome 17 open reading frame 57
|
|
chrX_-_102319096
|
1.050
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chr19_+_5681016
|
1.048
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr22_-_39268230
|
1.046
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr14_-_81687574
|
1.038
|
NM_201595
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa
|
|
chr3_+_51428720
|
1.037
|
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr15_+_44084039
|
1.034
|
NM_001018108
NM_001199875
NM_001199876
|
SERF2
|
small EDRK-rich factor 2
|
|
chrX_+_153686620
|
1.033
|
NM_017514
|
PLXNA3
|
plexin A3
|
|
chr17_-_74722843
|
1.024
|
NM_001081461
NM_015167
|
JMJD6
|
jumonji domain containing 6
|
|
chr17_-_46667596
|
1.018
|
|
HOXB3
|
homeobox B3
|
|
chr19_-_47734215
|
1.018
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr19_-_36545298
|
1.018
|
|
THAP8
|
THAP domain containing 8
|
|
chrX_+_100805415
|
1.017
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr5_-_132299191
|
1.016
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr2_-_26101301
|
1.011
|
NM_018263
|
ASXL2
|
additional sex combs like 2 (Drosophila)
|
|
chr19_+_1067536
|
1.005
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr17_-_74722614
|
1.002
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr16_+_81812899
|
1.002
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr19_+_4969123
|
0.995
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr11_-_85780105
|
0.994
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr16_+_87425998
|
0.991
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr4_-_84376943
|
0.989
|
NM_133636
|
HELQ
|
helicase, POLQ-like
|
|
chr16_+_81812897
|
0.981
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr17_-_74722629
|
0.974
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr12_-_123380628
|
0.974
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr9_-_19127494
|
0.973
|
NM_001122
|
PLIN2
|
perilipin 2
|
|
chr3_-_49823953
|
0.972
|
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr15_-_40213087
|
0.971
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr14_-_35591151
|
0.967
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr19_-_14606939
|
0.965
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr2_+_239047362
|
0.964
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr19_-_3985460
|
0.963
|
NM_001961
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr17_+_62503079
|
0.959
|
NM_138363
|
CEP95
|
centrosomal protein 95kDa
|
|
chr5_-_132298973
|
0.957
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr14_+_103388992
|
0.955
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr11_-_85779821
|
0.953
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|