|
chr12_+_57610577
|
4.002
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr19_+_16435632
|
3.781
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr12_+_125478151
|
3.179
|
NM_080626
|
BRI3BP
|
BRI3 binding protein
|
|
chr16_+_29817426
|
3.135
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr13_+_100633977
|
3.035
|
NM_007129
|
ZIC2
|
Zic family member 2
|
|
chr18_-_5296013
|
2.975
|
NM_001143823
NM_003409
|
ZFP161
|
zinc finger protein 161 homolog (mouse)
|
|
chr3_-_47620186
|
2.897
|
NM_001206943
NM_001206944
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr7_+_98246595
|
2.896
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr16_+_67465019
|
2.815
|
NM_000196
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr1_+_26798961
|
2.808
|
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr3_+_49507470
|
2.762
|
NM_001165928
NM_001177634
NM_001177635
NM_001177636
NM_001177637
NM_001177638
NM_001177639
NM_001177640
NM_001177641
NM_001177642
NM_001177644
NM_004393
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr2_-_215674292
|
2.755
|
NM_000465
|
BARD1
|
BRCA1 associated RING domain 1
|
|
chrX_-_17879355
|
2.688
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr15_+_41786055
|
2.669
|
NM_002220
|
ITPKA
|
inositol-trisphosphate 3-kinase A
|
|
chr12_+_70759974
|
2.668
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr3_+_110790460
|
2.602
|
NM_001243286
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr11_+_76092336
|
2.581
|
|
LOC100506127
|
putative uncharacterized protein FLJ37770-like
|
|
chr17_-_35293920
|
2.519
|
|
|
|
|
chr1_-_99470177
|
2.509
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr6_+_27100784
|
2.442
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr12_+_53846086
|
2.428
|
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr11_-_125933186
|
2.384
|
NM_001243597
NM_016952
|
CDON
|
Cdon homolog (mouse)
|
|
chr16_+_29817810
|
2.330
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr4_-_25032306
|
2.327
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr19_+_16435734
|
2.304
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr19_+_639878
|
2.292
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chr7_-_27239724
|
2.180
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr3_-_52090090
|
2.129
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr3_-_184870627
|
2.122
|
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr14_+_75894919
|
2.084
|
NM_001135048
NM_130469
|
JDP2
|
Jun dimerization protein 2
|
|
chr19_+_30302861
|
2.084
|
NM_001238
|
CCNE1
|
cyclin E1
|
|
chr3_+_110790774
|
2.058
|
NM_001243288
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr3_+_110790662
|
2.035
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr13_-_101327027
|
2.034
|
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr1_+_184356227
|
2.010
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr20_-_5100443
|
1.995
|
NM_182649
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr6_+_111408780
|
1.986
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr20_+_42086468
|
1.981
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr2_+_121103670
|
1.968
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr1_+_117452359
|
1.954
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr1_+_184356186
|
1.943
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr9_+_108006892
|
1.936
|
NM_080546
|
SLC44A1
|
solute carrier family 44, member 1
|
|
chr13_-_37494370
|
1.922
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr1_+_33722158
|
1.921
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr12_+_120554692
|
1.917
|
|
|
|
|
chr1_+_87797301
|
1.896
|
|
|
|
|
chr14_+_105941061
|
1.888
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr9_+_108006990
|
1.884
|
|
SLC44A1
|
solute carrier family 44, member 1
|
|
chr1_+_64239566
|
1.884
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr5_+_36152221
|
1.871
|
|
SKP2
|
S-phase kinase-associated protein 2 (p45)
|
|
chr6_+_34205029
|
1.864
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr7_+_2559396
|
1.848
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr10_+_82214074
|
1.822
|
|
TSPAN14
|
tetraspanin 14
|
|
chr17_+_40913233
|
1.821
|
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr2_+_16080639
|
1.817
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr5_+_36152142
|
1.816
|
NM_001243120
NM_005983
NM_032637
|
SKP2
|
S-phase kinase-associated protein 2 (p45)
|
|
chr15_+_96873845
|
1.806
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr11_-_19263144
|
1.798
|
|
E2F8
|
E2F transcription factor 8
|
|
chr1_+_184356128
|
1.778
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr1_-_6295967
|
1.778
|
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr9_+_126773851
|
1.770
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr17_+_14204326
|
1.763
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr10_+_82214071
|
1.763
|
|
TSPAN14
|
tetraspanin 14
|
|
chr2_-_86564632
|
1.756
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr6_-_42981656
|
1.733
|
|
MEA1
|
male-enhanced antigen 1
|
|
chr18_-_5295606
|
1.725
|
NM_001243702
|
ZFP161
|
zinc finger protein 161 homolog (mouse)
|
|
chr1_+_178995055
|
1.725
|
NM_014864
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr1_-_6295989
|
1.723
|
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chr1_+_178995175
|
1.722
|
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr3_+_37903643
|
1.720
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr18_-_72921046
|
1.705
|
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr9_+_116638544
|
1.704
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr2_+_181845368
|
1.704
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chrX_-_50213589
|
1.703
|
NM_001013742
|
DGKK
|
diacylglycerol kinase, kappa
|
|
chr10_-_13390018
|
1.693
|
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr15_-_77924730
|
1.684
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr1_-_32801606
|
1.670
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr20_-_60982334
|
1.665
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr14_-_38064440
|
1.635
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr6_+_34204928
|
1.629
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_32403975
|
1.626
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr5_+_52776263
|
1.624
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr19_-_48673559
|
1.621
|
NM_000234
|
LIG1
|
ligase I, DNA, ATP-dependent
|
|
chr6_+_135502521
|
1.620
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr6_+_35182182
|
1.620
|
NM_152753
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr14_+_29236882
|
1.615
|
|
FOXG1
|
forkhead box G1
|
|
chr11_+_66045645
|
1.602
|
NM_182553
|
CNIH2
|
cornichon homolog 2 (Drosophila)
|
|
chr12_+_66218568
|
1.595
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr1_-_212003576
|
1.591
|
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr15_-_59041812
|
1.589
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr18_-_51750460
|
1.586
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr14_+_94640648
|
1.579
|
NM_020958
NM_058237
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4
|
|
chr1_+_43148187
|
1.570
|
|
YBX1
|
Y box binding protein 1
|
|
chr9_+_108006937
|
1.563
|
|
SLC44A1
|
solute carrier family 44, member 1
|
|
chr2_+_181845088
|
1.552
|
NM_182678
NM_006357
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr10_+_82214066
|
1.536
|
|
TSPAN14
|
tetraspanin 14
|
|
chr10_+_70587293
|
1.525
|
NM_001130159
NM_001130160
NM_001130161
NM_152709
|
STOX1
|
storkhead box 1
|
|
chr6_+_35181721
|
1.523
|
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr12_+_125478234
|
1.518
|
|
BRI3BP
|
BRI3 binding protein
|
|
chr12_+_56137063
|
1.510
|
NM_005811
|
GDF11
|
growth differentiation factor 11
|
|
chr12_+_48166929
|
1.501
|
NM_017842
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1
|
|
chr1_-_205719078
|
1.501
|
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr1_-_23670743
|
1.494
|
NM_001102397
NM_001102398
NM_001102399
NM_005826
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr1_+_184356364
|
1.493
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr22_+_33197678
|
1.491
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr22_+_50609159
|
1.488
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr6_+_31126114
|
1.485
|
NM_001077511
NM_007109
|
TCF19
|
transcription factor 19
|
|
chr6_+_35227300
|
1.482
|
|
ZNF76
|
zinc finger protein 76
|
|
chr15_+_66995414
|
1.474
|
|
SMAD6
|
SMAD family member 6
|
|
chrX_+_18443702
|
1.467
|
NM_003159
|
CDKL5
|
cyclin-dependent kinase-like 5
|
|
chr3_+_181429711
|
1.467
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr10_+_28966419
|
1.467
|
NM_012342
|
BAMBI
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis)
|
|
chr12_+_66218879
|
1.466
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr6_+_111408648
|
1.465
|
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chrX_-_125300079
|
1.464
|
NM_001013628
|
DCAF12L2
|
DDB1 and CUL4 associated factor 12-like 2
|
|
chr14_-_105261850
|
1.459
|
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr19_-_38746978
|
1.452
|
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr2_+_120189342
|
1.452
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr20_-_39946119
|
1.450
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr8_-_95907423
|
1.446
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr1_+_212208918
|
1.444
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr19_+_18530423
|
1.436
|
|
|
|
|
chr9_-_112083005
|
1.434
|
NM_018424
NM_019114
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chrX_-_16888442
|
1.433
|
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr3_-_149688638
|
1.432
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr12_+_53773924
|
1.432
|
NM_001251825
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chrX_-_54209639
|
1.430
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chr15_+_40763159
|
1.429
|
NM_130468
|
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14
|
|
chr1_-_1535446
|
1.427
|
NM_001242659
|
LOC643988
|
uncharacterized LOC643988
|
|
chr13_-_101327102
|
1.426
|
NM_001079669
NM_032813
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr15_-_50647048
|
1.425
|
|
FLJ10038
|
uncharacterized protein FLJ10038
|
|
chr5_+_52776408
|
1.425
|
|
FST
|
follistatin
|
|
chr1_+_38259458
|
1.416
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr10_-_44880452
|
1.414
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr4_-_5890142
|
1.412
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr12_+_90103470
|
1.406
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chr7_+_2671630
|
1.403
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chrX_-_16888175
|
1.393
|
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr5_+_52776479
|
1.391
|
|
FST
|
follistatin
|
|
chr20_+_42086533
|
1.389
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chrX_+_30671456
|
1.388
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr14_+_50087400
|
1.383
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr3_-_184870727
|
1.379
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr5_+_133450353
|
1.378
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr10_-_98592186
|
1.375
|
|
|
|
|
chr10_+_65281177
|
1.373
|
|
REEP3
|
receptor accessory protein 3
|
|
chr1_+_178995004
|
1.371
|
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr13_+_100634257
|
1.369
|
|
ZIC2
|
Zic family member 2
|
|
chr1_-_1850704
|
1.368
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr8_+_26435466
|
1.367
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr12_-_123756164
|
1.362
|
NM_004642
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1
|
|
chr20_-_44718517
|
1.358
|
|
NCOA5
|
nuclear receptor coactivator 5
|
|
chr1_+_43148142
|
1.357
|
|
YBX1
|
Y box binding protein 1
|
|
chr17_-_76921205
|
1.354
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr7_+_90893735
|
1.347
|
NM_003505
|
FZD1
|
frizzled family receptor 1
|
|
chr7_+_30174343
|
1.346
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr1_+_43148105
|
1.346
|
|
YBX1
|
Y box binding protein 1
|
|
chr15_-_59041777
|
1.337
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr12_+_98909414
|
1.336
|
|
TMPO
|
thymopoietin
|
|
chr4_-_146859960
|
1.336
|
|
|
|
|
chr1_-_223537400
|
1.334
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr6_+_135502445
|
1.329
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr17_-_66287256
|
1.328
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr10_+_129705359
|
1.326
|
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr13_+_114239047
|
1.320
|
NM_007111
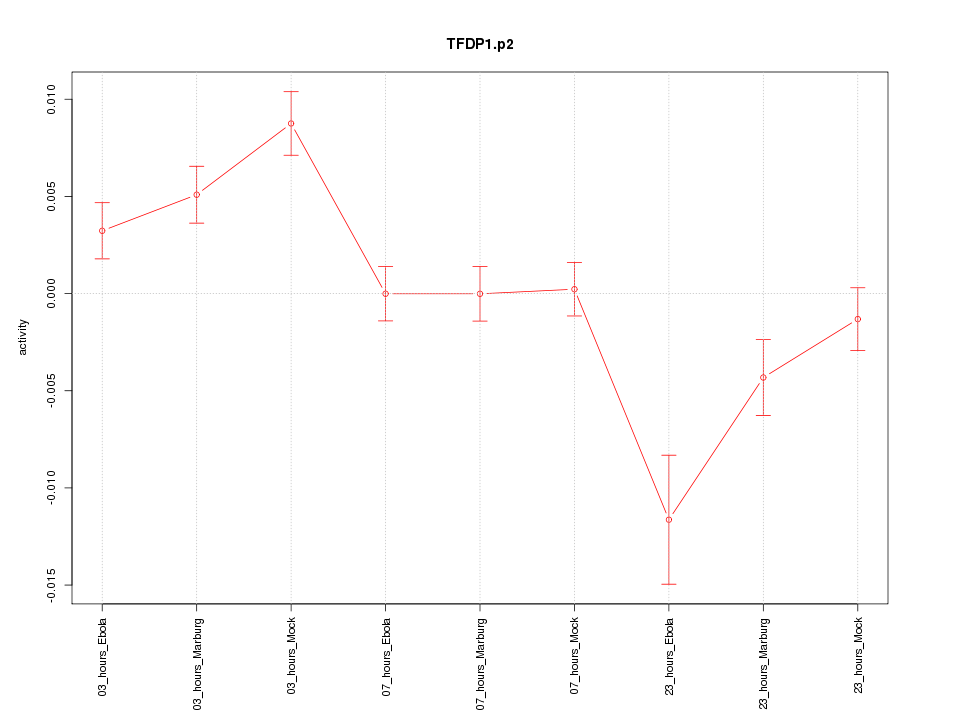
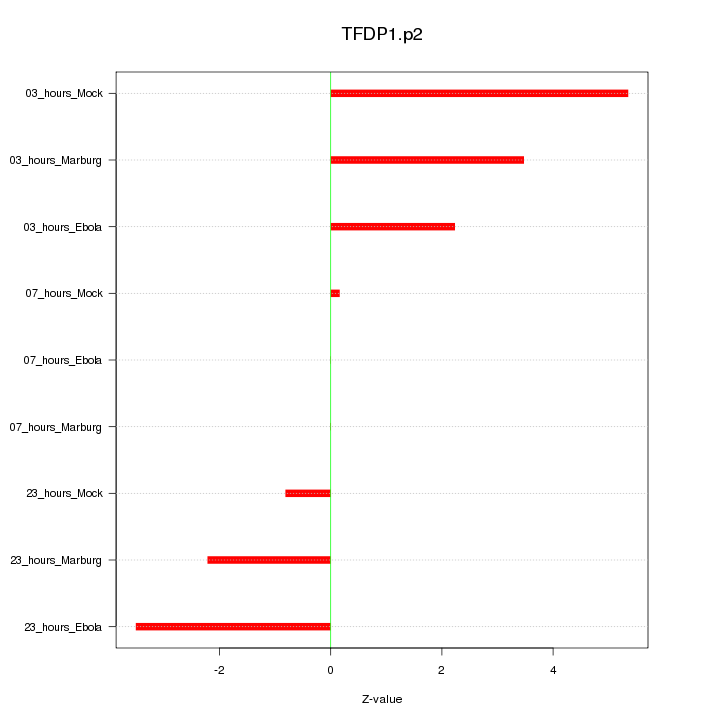
|
TFDP1
|
transcription factor Dp-1
|
|
chr2_-_165697927
|
1.316
|
NM_014900
|
COBLL1
|
COBL-like 1
|
|
chr17_-_1928177
|
1.315
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr2_+_150187028
|
1.314
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr10_+_70587562
|
1.307
|
NM_001130162
|
STOX1
|
storkhead box 1
|
|
chr7_+_139877154
|
1.304
|
|
LOC100134229
|
uncharacterized LOC100134229
|
|
chr6_-_107435635
|
1.302
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr6_+_71998508
|
1.295
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr2_-_43453724
|
1.295
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr14_-_38064176
|
1.292
|
|
FOXA1
|
forkhead box A1
|
|
chr12_+_50451486
|
1.290
|
NM_001095
NM_020039
|
ACCN2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr17_+_34890796
|
1.276
|
|
|
|
|
chr15_-_55880508
|
1.275
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr3_-_120068077
|
1.275
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr12_+_109535377
|
1.274
|
NM_080911
|
UNG
|
uracil-DNA glycosylase
|
|
chr1_+_183605207
|
1.269
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr12_-_21927614
|
1.264
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr10_+_82213892
|
1.263
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr15_+_59730272
|
1.262
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr1_+_26438267
|
1.262
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr1_+_43148168
|
1.261
|
|
YBX1
|
Y box binding protein 1
|
|
chr9_+_129089092
|
1.257
|
NM_001011703
NM_033446
|
FAM125B
|
family with sequence similarity 125, member B
|
|
chr15_+_90777463
|
1.257
|
NM_001013657
|
C15orf58
|
chromosome 15 open reading frame 58
|
|
chr4_+_6271558
|
1.255
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr5_+_176560627
|
1.253
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr9_+_128509616
|
1.253
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr3_-_113415313
|
1.248
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chrX_-_16888462
|
1.248
|
NM_002893
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr10_-_88854595
|
1.247
|
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr19_-_14201230
|
1.246
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chrX_-_16887974
|
1.245
|
NM_001198719
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr7_+_2671582
|
1.244
|
NM_025250
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr10_+_94449678
|
1.242
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr1_-_53793743
|
1.237
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr15_-_101792125
|
1.235
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr19_-_14228362
|
1.230
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|