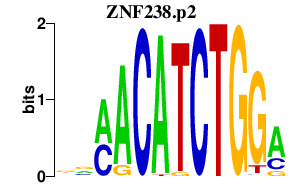
|
chr1_+_212781969
|
0.717
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr11_-_1606480
|
0.696
|
NM_001005922
|
KRTAP5-1
|
keratin associated protein 5-1
|
|
chr1_+_212782103
|
0.678
|
|
ATF3
|
activating transcription factor 3
|
|
chr17_+_48638339
|
0.576
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr17_+_47653474
|
0.485
|
|
NXPH3
|
neurexophilin 3
|
|
chr11_+_5474637
|
0.441
|
NM_001004754
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2
|
|
chr1_-_144520918
|
0.411
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr4_+_153864134
|
0.380
|
NM_033393
|
FHDC1
|
FH2 domain containing 1
|
|
chr1_-_147931877
|
0.353
|
|
FLJ39739
|
uncharacterized FLJ39739
|
|
chr11_-_5537815
|
0.338
|
NM_145053
|
UBQLNL
|
ubiquilin-like
|
|
chr7_+_28475233
|
0.338
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr13_+_97874704
|
0.337
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr13_+_97928196
|
0.336
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr12_+_27619742
|
0.334
|
NM_001145010
|
C12orf70
|
chromosome 12 open reading frame 70
|
|
chr8_-_8239188
|
0.330
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr6_-_163148800
|
0.328
|
NM_004562
NM_013987
NM_013988
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr11_+_71259465
|
0.323
|
NM_005553
|
KRTAP5-9
|
keratin associated protein 5-9
|
|
chr11_-_123065836
|
0.305
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr22_-_50708720
|
0.305
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr1_-_144520987
|
0.301
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr11_+_5775922
|
0.299
|
NM_001005175
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4
|
|
chr12_-_104443914
|
0.297
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr17_+_17398334
|
0.295
|
|
|
|
|
chr13_+_97874540
|
0.285
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr19_-_36001336
|
0.282
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr17_+_47653176
|
0.281
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr11_+_6220375
|
0.269
|
NM_001005178
|
OR52W1
|
olfactory receptor, family 52, subfamily W, member 1
|
|
chr15_+_40331417
|
0.268
|
|
LOC100131089
|
uncharacterized LOC100131089
|
|
chr1_+_6268919
|
0.267
|
|
RNF207
|
ring finger protein 207
|
|
chr2_+_108994366
|
0.259
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr11_-_1619481
|
0.252
|
NM_001004325
|
KRTAP5-2
|
keratin associated protein 5-2
|
|
chr1_+_26737306
|
0.247
|
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr1_-_150738292
|
0.246
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr19_+_3585541
|
0.244
|
NM_133261
|
GIPC3
|
GIPC PDZ domain containing family, member 3
|
|
chr1_-_33813261
|
0.240
|
|
|
|
|
chr8_-_145770680
|
0.240
|
|
ARHGAP39
|
Rho GTPase activating protein 39
|
|
chr16_-_30394133
|
0.235
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr3_-_114102488
|
0.231
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr13_+_97928414
|
0.227
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_-_33722285
|
0.226
|
NM_001166692
|
C11orf91
|
chromosome 11 open reading frame 91
|
|
chr16_+_50300461
|
0.225
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr1_-_247275527
|
0.223
|
NM_207401
|
C1orf229
|
chromosome 1 open reading frame 229
|
|
chr5_+_172068188
|
0.222
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr11_+_65405538
|
0.213
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr11_-_5863126
|
0.212
|
NM_001005167
|
OR52E6
|
olfactory receptor, family 52, subfamily E, member 6
|
|
chr16_-_30393856
|
0.208
|
|
SEPT1
|
septin 1
|
|
chr12_-_104443698
|
0.200
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr11_+_130029773
|
0.199
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr7_+_120969089
|
0.197
|
NM_057168
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr7_-_141541220
|
0.189
|
NM_001008270
NM_001171951
|
PRSS37
|
protease, serine, 37
|
|
chr19_-_44174497
|
0.188
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chrX_+_99899388
|
0.184
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr3_-_111314181
|
0.182
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr19_+_11649578
|
0.181
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr5_-_148758787
|
0.177
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr11_-_63684570
|
0.172
|
|
RCOR2
|
REST corepressor 2
|
|
chr6_+_160542862
|
0.170
|
NM_003057
NM_153187
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1
|
|
chr12_-_11175169
|
0.170
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr11_+_64323089
|
0.169
|
NM_018484
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11
|
|
chr8_+_81398416
|
0.167
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr12_-_58160823
|
0.164
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr17_+_45908992
|
0.161
|
NM_033413
|
LRRC46
|
leucine rich repeat containing 46
|
|
chr19_-_42636559
|
0.160
|
NM_001207025
NM_001207026
NM_001247994
NM_002698
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr15_-_43559054
|
0.160
|
NM_004245
NM_201631
|
TGM5
|
transglutaminase 5
|
|
chr7_+_90225607
|
0.159
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr5_-_151304322
|
0.156
|
NM_000171
NM_001146040
|
GLRA1
|
glycine receptor, alpha 1
|
|
chr7_+_127233688
|
0.156
|
NM_020369
|
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus)
|
|
chrX_+_99899162
|
0.156
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr16_+_29973341
|
0.154
|
NM_001083613
NM_194280
|
TMEM219
|
transmembrane protein 219
|
|
chr15_+_57210972
|
0.154
|
|
TCF12
|
transcription factor 12
|
|
chr15_+_57210889
|
0.152
|
|
TCF12
|
transcription factor 12
|
|
chr1_+_186798031
|
0.150
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr3_-_121379701
|
0.148
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr4_+_166794409
|
0.147
|
NM_001204760
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr17_+_42385780
|
0.146
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr9_+_87283453
|
0.144
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr7_-_124405680
|
0.143
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr22_+_38071612
|
0.139
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr16_+_57576576
|
0.138
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr12_+_104850986
|
0.137
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr19_+_54371170
|
0.135
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr11_-_83984230
|
0.133
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr7_+_102073953
|
0.131
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr19_+_54371150
|
0.130
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr10_+_35894337
|
0.129
|
NM_153368
|
GJD4
|
gap junction protein, delta 4, 40.1kDa
|
|
chr11_-_3692544
|
0.128
|
NM_020402
|
CHRNA10
|
cholinergic receptor, nicotinic, alpha 10
|
|
chr19_+_54371125
|
0.126
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr6_-_163148798
|
0.126
|
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr12_-_48597074
|
0.125
|
NM_001004134
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1
|
|
chr6_-_36807734
|
0.123
|
|
CPNE5
|
copine V
|
|
chr15_+_57210823
|
0.123
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr7_+_5013618
|
0.123
|
|
RNF216P1
|
ring finger protein 216 pseudogene 1
|
|
chr15_+_57210852
|
0.122
|
|
TCF12
|
transcription factor 12
|
|
chr2_+_130737234
|
0.122
|
NM_032144
|
RAB6C
|
RAB6C, member RAS oncogene family
|
|
chr9_-_99329092
|
0.122
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr18_+_9136742
|
0.121
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr17_-_74497406
|
0.120
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr4_+_15376174
|
0.119
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr19_+_9251052
|
0.118
|
NM_001190791
NM_020933
|
ZNF317
|
zinc finger protein 317
|
|
chr10_-_95241943
|
0.116
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr1_-_153931096
|
0.115
|
NM_181715
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr19_+_9486991
|
0.115
|
NM_001172651
|
ZNF177
|
zinc finger protein 177
|
|
chr20_+_30467596
|
0.115
|
|
TTLL9
|
tubulin tyrosine ligase-like family, member 9
|
|
chr7_+_102074002
|
0.114
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr18_+_3412071
|
0.114
|
NM_174886
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr12_-_15942116
|
0.113
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr5_-_134369841
|
0.113
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr7_-_44105162
|
0.112
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr8_-_125384916
|
0.112
|
NM_194291
|
TMEM65
|
transmembrane protein 65
|
|
chr5_+_15500539
|
0.111
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr13_-_60587298
|
0.111
|
NM_030932
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr1_+_19934573
|
0.110
|
|
|
|
|
chr5_-_39425030
|
0.110
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr19_+_48673936
|
0.110
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr19_+_56153417
|
0.109
|
NM_016202
NM_001163423
|
ZNF580
|
zinc finger protein 580
|
|
chr10_+_81073763
|
0.109
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr20_-_48770252
|
0.107
|
|
TMEM189
|
transmembrane protein 189
|
|
chr6_-_163148759
|
0.107
|
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr3_+_100211408
|
0.105
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr6_-_16761600
|
0.104
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr11_+_10326618
|
0.102
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr1_-_153930911
|
0.102
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr20_-_48770227
|
0.102
|
|
TMEM189
|
transmembrane protein 189
|
|
chr16_+_57576536
|
0.099
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr17_-_34195745
|
0.099
|
NM_152781
|
C17orf66
|
chromosome 17 open reading frame 66
|
|
chr10_-_22292640
|
0.099
|
NM_022365
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr10_-_22292362
|
0.098
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr2_-_225378320
|
0.098
|
|
CUL3
|
cullin 3
|
|
chr10_-_103543156
|
0.098
|
NM_006993
|
NPM3
|
nucleophosmin/nucleoplasmin 3
|
|
chr19_+_49617631
|
0.097
|
|
LIN7B
|
lin-7 homolog B (C. elegans)
|
|
chr12_-_50222144
|
0.097
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chr1_-_153931077
|
0.096
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr3_+_100211554
|
0.096
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr2_-_211018294
|
0.096
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr3_+_50316517
|
0.095
|
NM_153215
|
C3orf45
|
chromosome 3 open reading frame 45
|
|
chr11_-_11643542
|
0.094
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr1_-_156647088
|
0.094
|
NM_006617
|
NES
|
nestin
|
|
chr1_-_100643770
|
0.093
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr19_+_18668571
|
0.093
|
NM_001171948
NM_001171949
NM_024069
|
KXD1
|
KxDL motif containing 1
|
|
chr10_+_116853118
|
0.091
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr12_-_48119224
|
0.091
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr15_+_78556485
|
0.090
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr16_+_28891234
|
0.090
|
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr5_+_15500288
|
0.088
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr12_+_104850762
|
0.087
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr2_+_113279582
|
0.087
|
|
TTL
|
tubulin tyrosine ligase
|
|
chr10_-_3827205
|
0.087
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr11_-_34379526
|
0.086
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr4_-_105416050
|
0.086
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr6_-_32191779
|
0.085
|
NM_004557
|
NOTCH4
|
notch 4
|
|
chr11_-_63684315
|
0.084
|
NM_173587
|
RCOR2
|
REST corepressor 2
|
|
chrX_+_100333835
|
0.084
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr12_+_104850784
|
0.084
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr10_-_103543148
|
0.084
|
|
NPM3
|
nucleophosmin/nucleoplasmin 3
|
|
chr16_-_31146992
|
0.083
|
|
PRSS8
|
protease, serine, 8
|
|
chr2_+_29038862
|
0.083
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr12_-_108154924
|
0.082
|
|
PRDM4
|
PR domain containing 4
|
|
chr16_+_4838397
|
0.082
|
NM_001253790
NM_001253791
|
LOC440335
|
uncharacterized LOC440335
|
|
chr6_-_128222175
|
0.081
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr13_+_113698907
|
0.081
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr12_+_104850641
|
0.081
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr17_+_64960754
|
0.080
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr10_+_31607419
|
0.078
|
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr6_+_33239855
|
0.075
|
|
RPS18
|
ribosomal protein S18
|
|
chr17_-_4806368
|
0.075
|
NM_000080
|
CHRNE
|
cholinergic receptor, nicotinic, epsilon
|
|
chr5_+_49962920
|
0.075
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr19_+_49617611
|
0.074
|
NM_022165
|
LIN7B
|
lin-7 homolog B (C. elegans)
|
|
chr2_+_130939750
|
0.074
|
|
MZT2B
|
mitotic spindle organizing protein 2B
|
|
chr5_-_39424930
|
0.074
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr10_+_45406629
|
0.073
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr19_+_18668652
|
0.073
|
|
KXD1
|
KxDL motif containing 1
|
|
chr6_+_42714686
|
0.073
|
|
KIAA0240
|
KIAA0240
|
|
chr1_-_217263195
|
0.072
|
NM_001243505
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_-_64777703
|
0.072
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr9_+_103340335
|
0.071
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr9_+_100174369
|
0.071
|
|
TDRD7
|
tudor domain containing 7
|
|
chr2_+_219187858
|
0.071
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr11_+_76494132
|
0.071
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr20_-_48770302
|
0.071
|
NM_001162505
NM_199129
NM_199203
|
TMEM189
TMEM189-UBE2V1
|
transmembrane protein 189
TMEM189-UBE2V1 readthrough
|
|
chrX_+_118108575
|
0.071
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr2_+_219187975
|
0.071
|
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr6_-_53530505
|
0.070
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr2_+_33359607
|
0.070
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr19_-_35625610
|
0.070
|
|
LGI4
|
leucine-rich repeat LGI family, member 4
|
|
chr7_-_37025696
|
0.070
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr4_-_82126200
|
0.069
|
NM_006259
|
PRKG2
|
protein kinase, cGMP-dependent, type II
|
|
chr3_-_121379757
|
0.069
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr12_-_52225700
|
0.069
|
NM_001013690
|
FIGNL2
|
fidgetin-like 2
|
|
chr22_-_23484208
|
0.068
|
NM_014433
|
RTDR1
|
rhabdoid tumor deletion region gene 1
|
|
chr9_+_134305476
|
0.068
|
NM_013318
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chrX_+_118109314
|
0.066
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr7_-_141673572
|
0.066
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr11_+_63655986
|
0.066
|
NM_017490
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr15_+_90319588
|
0.066
|
NM_001039958
|
MESP2
|
mesoderm posterior 2 homolog (mouse)
|
|
chr12_-_15942350
|
0.066
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr12_+_26348488
|
0.065
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr6_-_138189313
|
0.065
|
|
|
|
|
chr6_+_33239798
|
0.065
|
NM_022551
|
RPS18
|
ribosomal protein S18
|
|
chr3_-_172858959
|
0.065
|
NM_031955
|
SPATA16
|
spermatogenesis associated 16
|
|
chr19_-_42463500
|
0.065
|
NM_006423
|
RABAC1
|
Rab acceptor 1 (prenylated)
|