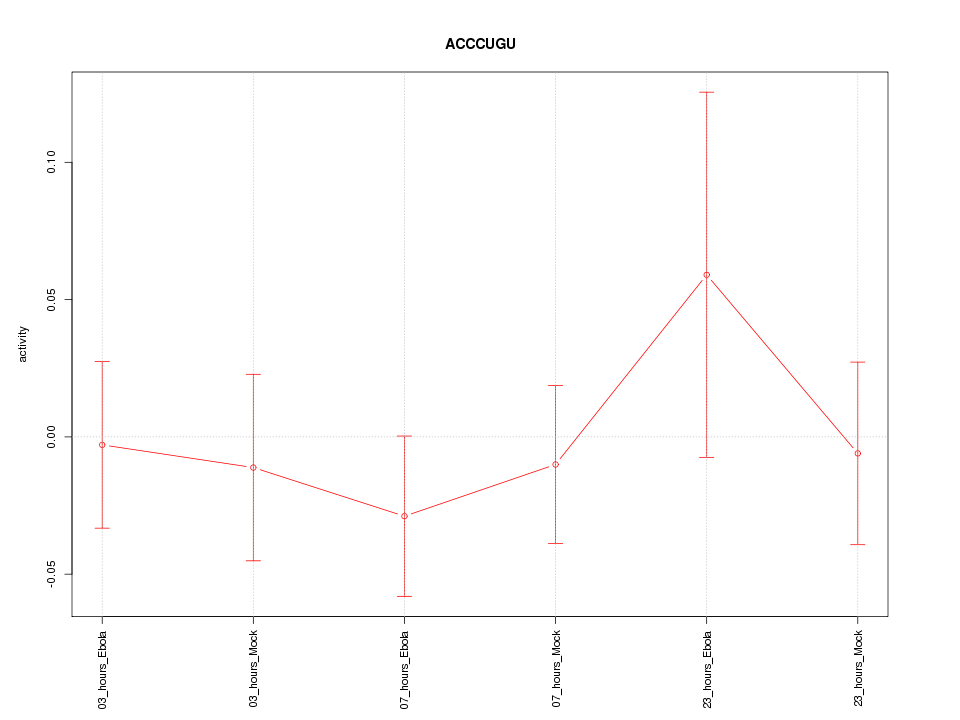
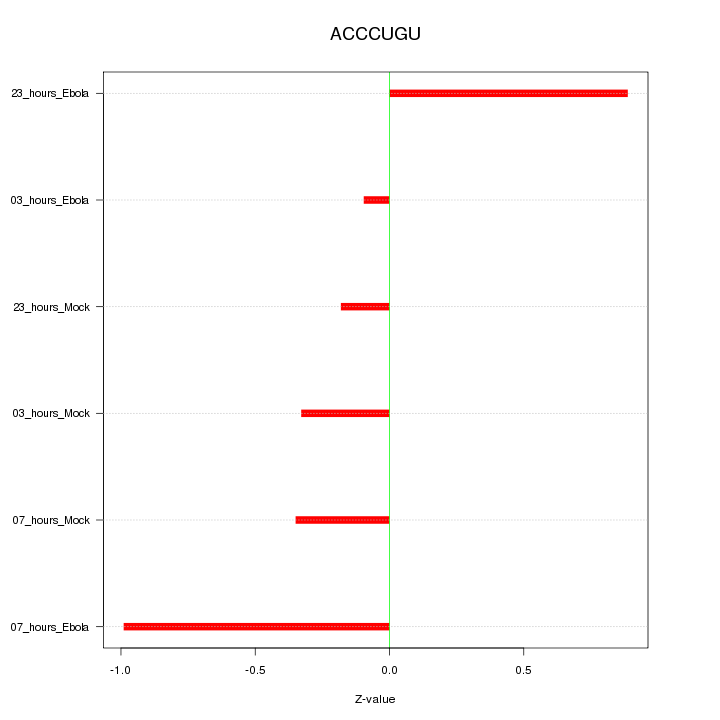
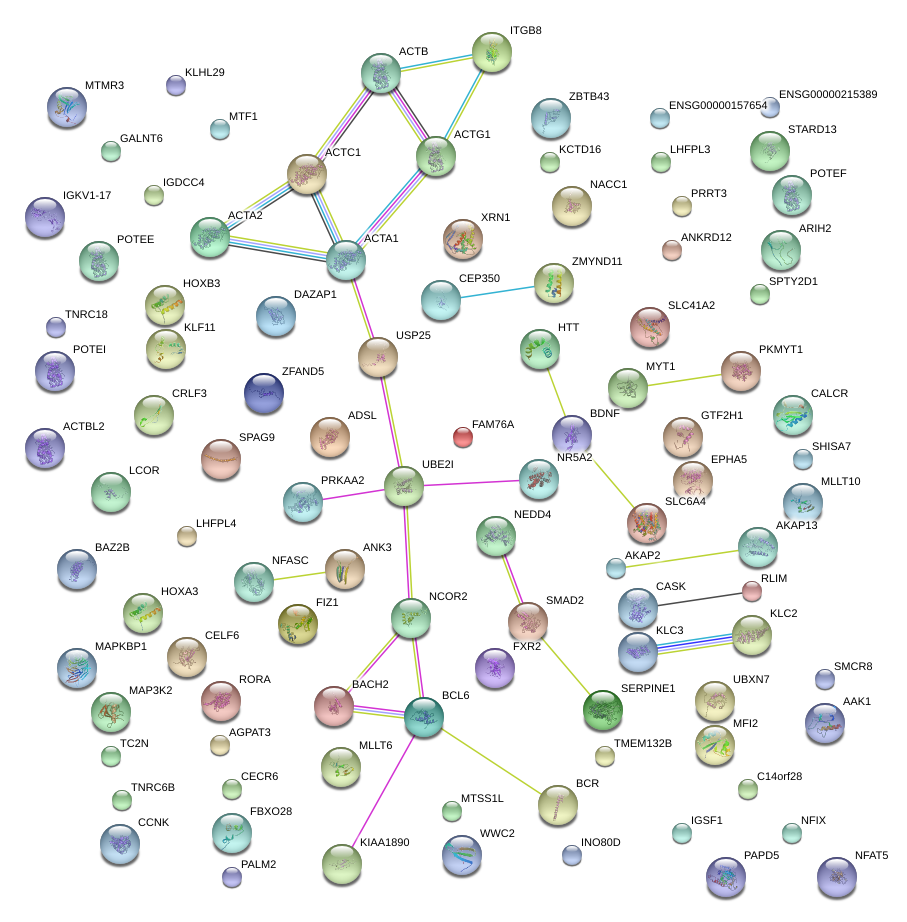
|
chr6_-_91006460
|
1.312
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_-_91006580
|
1.286
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr15_-_60919642
|
1.017
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_60884615
|
1.016
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_61521478
|
0.963
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr22_-_17602142
|
0.489
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr14_+_45366426
|
0.470
|
NM_001017923
|
C14orf28
|
chromosome 14 open reading frame 28
|
|
chr2_-_206950895
|
0.441
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr19_+_13105970
|
0.441
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr7_-_93204035
|
0.440
|
NM_001164737
NM_001742
|
CALCR
|
calcitonin receptor
|
|
chr17_-_7518124
|
0.431
|
NM_004860
|
FXR2
|
fragile X mental retardation, autosomal homolog 2
|
|
chr17_-_49198084
|
0.425
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr18_+_9137560
|
0.422
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr2_+_10184371
|
0.411
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr2_-_69870976
|
0.398
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr15_-_56285732
|
0.395
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr3_-_187452669
|
0.385
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_-_187454247
|
0.376
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr22_+_40573879
|
0.367
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr15_-_65715385
|
0.365
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr7_-_27159213
|
0.338
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr7_-_27166638
|
0.332
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr3_-_187463474
|
0.328
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr4_-_66535652
|
0.317
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr7_+_100081549
|
0.314
|
NM_173564
|
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1
|
|
chr2_+_10183630
|
0.313
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr16_+_69599868
|
0.292
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr18_+_9136742
|
0.273
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr11_+_66024746
|
0.266
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr12_-_125052009
|
0.263
|
NM_001077261
NM_001206654
NM_006312
|
NCOR2
|
nuclear receptor corepressor 2
|
|
chr3_-_9994057
|
0.261
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr22_+_23522551
|
0.260
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr22_+_40440664
|
0.258
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr11_+_66025070
|
0.232
|
NM_001134776
NM_001134774
NM_022822
|
KLC2
|
kinesin light chain 2
|
|
chr10_-_62332666
|
0.228
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_62149487
|
0.217
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr9_+_129567260
|
0.215
|
NM_001135776
NM_014007
|
ZBTB43
|
zinc finger and BTB domain containing 43
|
|
chr10_-_62493282
|
0.205
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr15_-_56209305
|
0.203
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr7_-_93189121
|
0.203
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chr17_+_18218569
|
0.185
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr17_-_46651809
|
0.185
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr3_-_9595412
|
0.177
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr1_+_204913353
|
0.174
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr16_-_70719854
|
0.172
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr7_+_100770378
|
0.172
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr22_+_30279064
|
0.168
|
NM_021090
NM_153050
NM_153051
|
MTMR3
|
myotubularin related protein 3
|
|
chr10_+_181410
|
0.164
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr16_+_50186809
|
0.154
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr9_-_74979829
|
0.153
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr15_+_42066631
|
0.148
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr13_-_33760215
|
0.138
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr19_+_1407531
|
0.131
|
NM_018959
NM_170711
|
DAZAP1
|
DAZ associated protein 1
|
|
chr19_+_13229101
|
0.130
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr1_+_179923870
|
0.129
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr11_-_27681195
|
0.126
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_+_18343794
|
0.123
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr2_-_128100673
|
0.121
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr11_-_27722446
|
0.120
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_+_23608297
|
0.118
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr12_+_125811161
|
0.118
|
NM_052907
|
TMEM132B
|
transmembrane protein 132B
|
|
chr1_+_204797774
|
0.116
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr3_-_142166782
|
0.116
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr19_-_55954229
|
0.115
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chrX_-_73834460
|
0.111
|
NM_016120
NM_183353
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr10_+_98592658
|
0.107
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr10_+_180404
|
0.105
|
NM_006624
NM_001202465
NM_001202466
NM_212479
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr1_+_57110745
|
0.104
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr21_+_45345278
|
0.101
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr10_-_61900659
|
0.099
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_28052467
|
0.094
|
NM_001143912
NM_001143913
NM_001143914
NM_001143915
NM_152660
|
FAM76A
|
family with sequence similarity 76, member A
|
|
chr11_-_27721179
|
0.093
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_+_20370724
|
0.089
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr17_+_36861857
|
0.088
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr14_+_99947732
|
0.088
|
NM_001099402
|
CCNK
|
cyclin K
|
|
chr2_-_160472960
|
0.087
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr9_+_112403067
|
0.081
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr21_+_45285101
|
0.080
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr15_+_86220267
|
0.074
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr15_+_85923739
|
0.073
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr1_+_224301790
|
0.073
|
NM_001136115
NM_015176
|
FBXO28
|
F-box protein 28
|
|
chr7_-_106301329
|
0.073
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr17_-_29151724
|
0.070
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr7_-_5463176
|
0.069
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr5_+_143550395
|
0.069
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr11_-_18655962
|
0.068
|
NM_194285
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr12_-_105322056
|
0.066
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr1_-_38325224
|
0.064
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr13_-_33859835
|
0.063
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr15_-_72612247
|
0.063
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr1_+_199996729
|
0.061
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr19_-_56110851
|
0.059
|
NM_032836
|
FIZ1
|
FLT3-interacting zinc finger 1
|
|
chr11_-_27722599
|
0.059
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr4_+_184020343
|
0.057
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr17_-_79479831
|
0.055
|
NM_001199954
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr11_-_27741192
|
0.054
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_-_41782230
|
0.037
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr18_-_45456925
|
0.036
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr3_-_196159304
|
0.036
|
NM_015562
|
UBXN7
|
UBX domain protein 7
|
|
chr21_+_17102370
|
0.034
|
NM_013396
|
USP25
|
ubiquitin specific peptidase 25
|
|
chr16_+_1359622
|
0.034
|
NM_003345
NM_194259
|
UBE2I
|
ubiquitin-conjugating enzyme E2I
|
|
chr4_+_3076236
|
0.033
|
NM_002111
|
HTT
|
huntingtin
|
|
chr13_-_33780142
|
0.033
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_48956272
|
0.030
|
NM_006321
|
ARIH2
|
ariadne homolog 2 (Drosophila)
|
|
chr8_-_4852262
|
0.025
|
NM_033225
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chrX_-_130423387
|
0.024
|
NM_001170961
NM_001170962
NM_001555
NM_205833
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chrX_-_129402753
|
0.021
|
NM_017666
|
ZNF280C
|
zinc finger protein 280C
|
|
chr7_-_92465929
|
0.020
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr9_-_95527002
|
0.018
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chr10_-_88281515
|
0.018
|
NM_015045
|
WAPAL
|
wings apart-like homolog (Drosophila)
|
|
chr8_-_16050220
|
0.016
|
NM_002445
NM_138715
NM_138716
|
MSR1
|
macrophage scavenger receptor 1
|
|
chr11_-_27743604
|
0.016
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_+_71377471
|
0.015
|
NM_001044305
NM_021940
|
SMAP1
|
small ArfGAP 1
|
|
chr14_+_23775970
|
0.014
|
NM_001199864
NM_004050
NM_001199839
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough
BCL2-like 2
|
|
chr3_+_183873253
|
0.010
|
NM_004423
|
DVL3
|
dishevelled, dsh homolog 3 (Drosophila)
|
|
chr11_-_27721975
|
0.008
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr3_-_125239049
|
0.005
|
NM_003794
|
SNX4
|
sorting nexin 4
|
|
chr9_+_112542496
|
0.004
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr1_-_38230804
|
0.004
|
NM_001099439
NM_173641
|
EPHA10
|
EPH receptor A10
|
|
chr15_-_72598655
|
0.004
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr7_-_92463211
|
0.004
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|