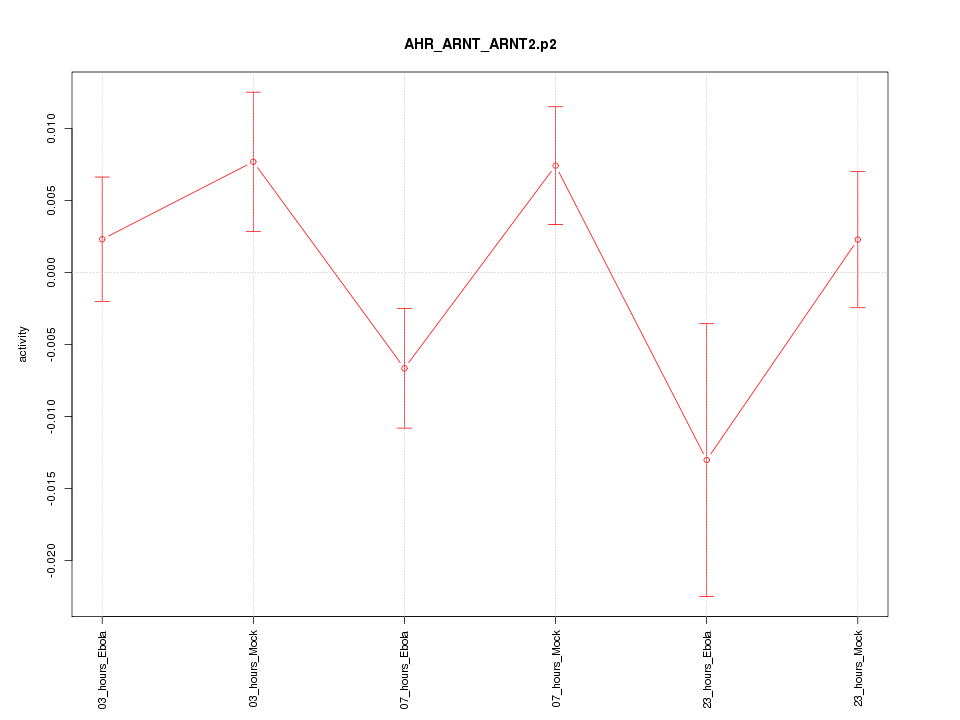
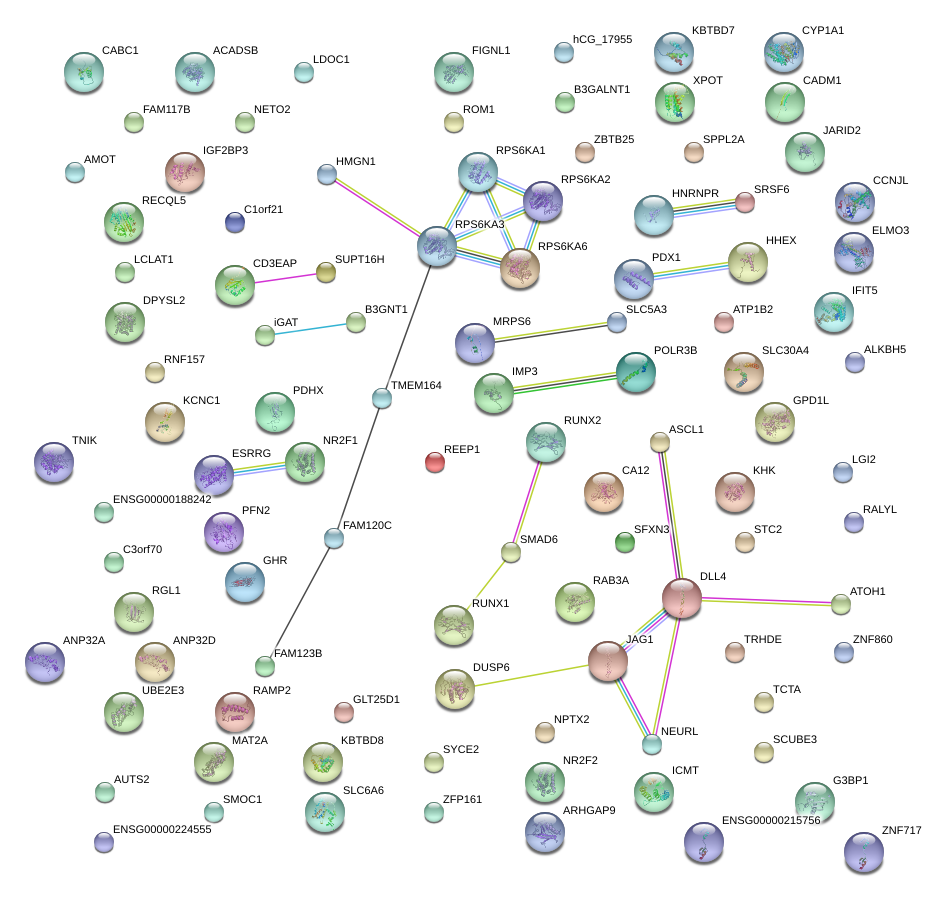
|
chr4_+_94750041
|
2.078
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chr1_+_109642814
|
2.005
|
|
SCARNA2
|
small Cajal body-specific RNA 2
|
|
chr19_-_18314731
|
1.676
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr2_+_203499819
|
1.111
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr10_-_98592186
|
1.035
|
|
|
|
|
chr5_+_151151516
|
1.023
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr5_+_151151514
|
0.974
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr5_+_151151483
|
0.957
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr4_-_25032306
|
0.855
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr5_-_159739482
|
0.855
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr5_+_151151511
|
0.841
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr13_-_41768392
|
0.826
|
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr5_+_151151462
|
0.815
|
NM_005754
NM_198395
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr3_-_184870727
|
0.814
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr2_+_203500211
|
0.807
|
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chrX_-_112084006
|
0.759
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr15_-_75932509
|
0.723
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr4_-_146859960
|
0.707
|
|
|
|
|
chr13_+_28494136
|
0.686
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr7_+_98246595
|
0.679
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr6_+_15245702
|
0.671
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr15_-_69113089
|
0.658
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chrX_+_109246350
|
0.656
|
|
TMEM164
|
transmembrane protein 164
|
|
chr18_-_5296013
|
0.627
|
NM_001143823
NM_003409
|
ZFP161
|
zinc finger protein 161 homolog (mouse)
|
|
chr2_+_85766291
|
0.610
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr3_-_149688638
|
0.604
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr10_+_124768498
|
0.597
|
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr15_-_69113213
|
0.589
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr19_-_13030074
|
0.588
|
NM_001105578
|
SYCE2
|
synaptonemal complex central element protein 2
|
|
chr10_+_124768401
|
0.583
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr15_+_66995414
|
0.560
|
|
SMAD6
|
SMAD family member 6
|
|
chr2_+_203499667
|
0.545
|
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr10_+_91174438
|
0.541
|
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5
|
|
chr3_+_52273247
|
0.539
|
|
|
|
|
chr10_+_105344714
|
0.518
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr1_-_23670743
|
0.515
|
NM_001102397
NM_001102398
NM_001102399
NM_005826
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R
|
|
chrX_+_109246338
|
0.514
|
NM_032227
|
TMEM164
|
transmembrane protein 164
|
|
chr8_+_26435339
|
0.511
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_160822625
|
0.509
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr10_+_102792166
|
0.506
|
|
SFXN3
|
sideroflexin 3
|
|
chr20_+_42086533
|
0.504
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr18_+_77659446
|
0.498
|
|
|
|
|
chr3_+_32023265
|
0.496
|
NM_001137674
|
ZNF860
|
zinc finger protein 860
|
|
chr19_+_17666457
|
0.495
|
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr7_+_139877154
|
0.493
|
|
LOC100134229
|
uncharacterized LOC100134229
|
|
chr17_-_74236272
|
0.493
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr5_-_172755418
|
0.489
|
|
STC2
|
stanniocalcin 2
|
|
chr5_-_172755195
|
0.487
|
|
STC2
|
stanniocalcin 2
|
|
chr5_+_42423811
|
0.477
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr7_-_50518021
|
0.475
|
NM_001042762
NM_022116
|
FIGNL1
|
fidgetin-like 1
|
|
chr11_-_66115016
|
0.466
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr11_+_62380212
|
0.465
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr21_-_40720842
|
0.461
|
|
HMGN1
|
high mobility group nucleosome binding domain 1
|
|
chr10_+_102792161
|
0.454
|
|
SFXN3
|
sideroflexin 3
|
|
chr17_+_18087091
|
0.452
|
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr19_+_663381
|
0.448
|
|
|
|
|
chr20_+_42086468
|
0.443
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr2_+_27310079
|
0.441
|
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr2_+_181845088
|
0.439
|
NM_182678
NM_006357
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr3_-_184870627
|
0.438
|
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr8_+_85095501
|
0.433
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr1_+_184356364
|
0.430
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr2_+_30670082
|
0.428
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr19_-_46220589
|
0.427
|
|
|
|
|
chr17_+_40913233
|
0.427
|
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr20_-_10654429
|
0.424
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr10_+_94449678
|
0.421
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr12_+_72666462
|
0.419
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr1_-_1535446
|
0.418
|
NM_001242659
|
LOC643988
|
uncharacterized LOC643988
|
|
chr17_-_56084498
|
0.416
|
|
|
|
|
chr19_+_45909466
|
0.415
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr11_-_575848
|
0.413
|
|
LOC143666
|
uncharacterized LOC143666
|
|
chr1_+_184356186
|
0.410
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr12_-_89745753
|
0.409
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr12_+_103351451
|
0.406
|
NM_004316
|
ASCL1
|
achaete-scute complex homolog 1 (Drosophila)
|
|
chr3_+_49449753
|
0.405
|
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr2_-_86564632
|
0.404
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr16_-_47177902
|
0.404
|
NM_001201477
NM_018092
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr14_-_21852085
|
0.403
|
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr1_-_216896640
|
0.400
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_+_35182182
|
0.396
|
NM_152753
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr3_-_75834661
|
0.391
|
|
ZNF717
|
zinc finger protein 717
|
|
chr7_+_70231163
|
0.391
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr14_-_64970319
|
0.390
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr16_-_47177832
|
0.390
|
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr11_+_76092336
|
0.387
|
|
LOC100506127
|
putative uncharacterized protein FLJ37770-like
|
|
chr14_+_70346113
|
0.387
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr15_-_63673918
|
0.386
|
|
CA12
|
carbonic anhydrase XII
|
|
chr16_-_87350947
|
0.385
|
NM_001195124
NM_001195125
|
C16orf95
|
chromosome 16 open reading frame 95
|
|
chr1_+_184356227
|
0.384
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr15_+_41221530
|
0.382
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr21_-_36260972
|
0.382
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_+_14444083
|
0.381
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr5_-_472916
|
0.377
|
|
PP7080
|
uncharacterized LOC25845
|
|
chr11_+_17757494
|
0.375
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr14_-_21852161
|
0.373
|
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr1_+_227127959
|
0.373
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr21_-_40720938
|
0.370
|
|
HMGN1
|
high mobility group nucleosome binding domain 1
|
|
chr17_+_7554711
|
0.370
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr19_+_17666502
|
0.370
|
NM_024656
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr21_+_35445808
|
0.369
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr12_+_64798050
|
0.369
|
NM_007235
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr3_+_67048726
|
0.367
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr15_-_69113255
|
0.367
|
NM_006305
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr1_-_6295989
|
0.366
|
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chrX_-_63425484
|
0.362
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr15_-_75017710
|
0.360
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chrX_-_20284708
|
0.359
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr19_+_17666381
|
0.358
|
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr3_+_32148136
|
0.357
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr3_-_171177851
|
0.356
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chrX_-_148622357
|
0.350
|
|
LOC100131434
|
uncharacterized LOC100131434
|
|
chr15_-_45815001
|
0.350
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr15_+_96873845
|
0.347
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_-_149688886
|
0.346
|
|
PFN2
|
profilin 2
|
|
chr12_+_106751550
|
0.346
|
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B
|
|
chr17_+_40913211
|
0.345
|
NM_005854
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chrX_-_54209639
|
0.345
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chr16_+_67233013
|
0.343
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chrX_-_140271275
|
0.341
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr1_+_183774214
|
0.341
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr3_+_32148000
|
0.339
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr11_-_115375065
|
0.336
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chrX_-_140271293
|
0.334
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr16_-_51185150
|
0.333
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr19_-_46088074
|
0.331
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr3_-_107809754
|
0.329
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr17_-_56084514
|
0.329
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr10_-_13390018
|
0.328
|
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr6_-_42419782
|
0.328
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr19_+_10527448
|
0.328
|
NM_001243121
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr8_+_26435466
|
0.327
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr1_+_227127993
|
0.326
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_+_142618798
|
0.324
|
|
|
|
|
chr17_+_54671059
|
0.324
|
NM_005450
|
NOG
|
noggin
|
|
chr2_+_181845368
|
0.322
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr12_-_21927614
|
0.322
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr1_-_205649616
|
0.322
|
NM_033102
|
SLC45A3
|
solute carrier family 45, member 3
|
|
chr5_+_134181729
|
0.322
|
|
C5orf24
|
chromosome 5 open reading frame 24
|
|
chr19_-_2427530
|
0.322
|
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr15_+_42565784
|
0.319
|
|
GANC
|
glucosidase, alpha; neutral C
|
|
chr17_+_18086813
|
0.317
|
NM_017758
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr20_-_10654139
|
0.317
|
|
JAG1
|
jagged 1
|
|
chr2_+_101179395
|
0.316
|
NM_024065
|
PDCL3
|
phosducin-like 3
|
|
chr17_+_28256873
|
0.315
|
NM_001145053
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr1_-_54304107
|
0.314
|
NM_001168551
NM_018087
|
TMEM48
|
transmembrane protein 48
|
|
chr14_+_103800678
|
0.311
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr7_+_94285736
|
0.311
|
|
PEG10
|
paternally expressed 10
|
|
chr7_-_93519990
|
0.307
|
NM_006528
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chrX_-_154493736
|
0.307
|
NM_171998
|
RAB39B
|
RAB39B, member RAS oncogene family
|
|
chr5_+_6633531
|
0.305
|
|
SRD5A1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1)
|
|
chr16_+_15489586
|
0.303
|
NM_001128423
NM_173803
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr10_-_13390189
|
0.302
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr20_-_3869713
|
0.301
|
|
|
|
|
chr1_+_227127999
|
0.300
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr5_-_83680191
|
0.299
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr13_-_41768533
|
0.299
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr13_+_100634257
|
0.299
|
|
ZIC2
|
Zic family member 2
|
|
chr1_+_184356128
|
0.298
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr9_-_134151905
|
0.298
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr10_-_21805715
|
0.296
|
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr17_-_56084611
|
0.296
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chrX_+_150565674
|
0.295
|
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae)
|
|
chr5_-_137089434
|
0.295
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr7_+_94285676
|
0.294
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr1_-_54303933
|
0.294
|
|
TMEM48
|
transmembrane protein 48
|
|
chr12_-_16758839
|
0.291
|
NM_001243612
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_-_38746978
|
0.290
|
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr6_+_99282579
|
0.290
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr6_-_167369990
|
0.289
|
|
RNASET2
|
ribonuclease T2
|
|
chr16_-_31470582
|
0.289
|
|
|
|
|
chr17_+_7210841
|
0.286
|
NM_001970
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr5_-_111093792
|
0.285
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_145718586
|
0.285
|
NM_002700
|
POU4F3
|
POU class 4 homeobox 3
|
|
chr2_+_27309610
|
0.284
|
NM_000221
NM_006488
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr5_-_36241684
|
0.283
|
|
NADKD1
|
NAD kinase domain containing 1
|
|
chr9_+_119449573
|
0.283
|
NM_001099679
NM_012210
|
TRIM32
|
tripartite motif containing 32
|
|
chrX_-_16888462
|
0.281
|
NM_002893
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr1_-_47184790
|
0.280
|
|
|
|
|
chr14_+_29236171
|
0.280
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chrX_+_109245851
|
0.277
|
NM_017698
|
TMEM164
|
transmembrane protein 164
|
|
chr7_-_84816170
|
0.277
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr9_+_80911990
|
0.277
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chrX_-_16888175
|
0.276
|
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr6_+_21593963
|
0.276
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr16_+_15489619
|
0.276
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr12_-_72057389
|
0.276
|
|
|
|
|
chr1_+_226736603
|
0.275
|
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr1_+_119683006
|
0.275
|
|
|
|
|
chr22_-_31885512
|
0.274
|
NM_001164501
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1
|
|
chr5_-_146258143
|
0.274
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr7_+_94285620
|
0.273
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr10_+_101089109
|
0.273
|
|
CNNM1
|
cyclin M1
|
|
chr17_+_34891340
|
0.272
|
NM_178517
|
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W
|
|
chr1_-_113498615
|
0.272
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chrX_-_16888442
|
0.272
|
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr19_+_4909447
|
0.271
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr13_+_114238978
|
0.270
|
|
TFDP1
|
transcription factor Dp-1
|
|
chr3_-_71632741
|
0.270
|
NM_001244808
NM_001012505
NM_001244810
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr6_+_15249034
|
0.270
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|