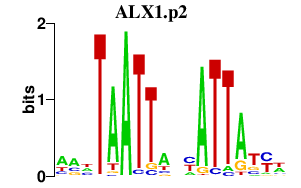
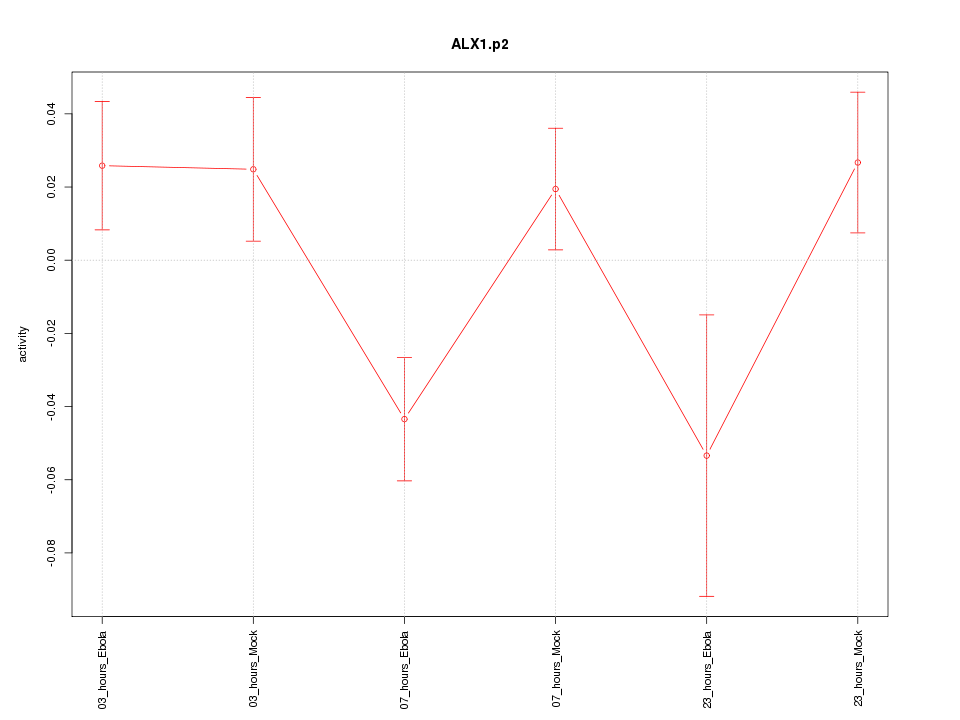
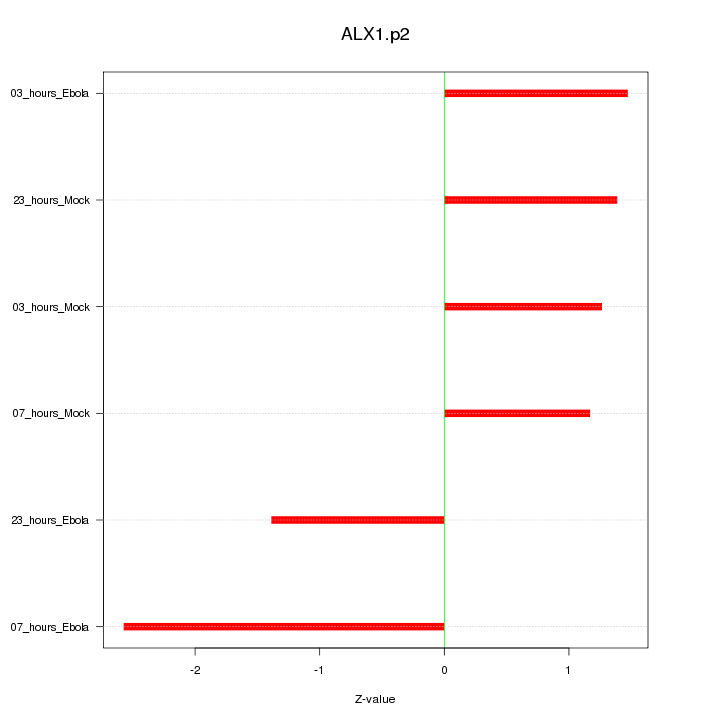
|
chr6_+_26251826
|
2.693
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr6_-_26189303
|
2.137
|
NM_003539
|
HIST1H4D
|
histone cluster 1, H4d
|
|
chr10_-_42863435
|
1.790
|
|
LOC441666
|
zinc finger protein 91 pseudogene
|
|
chr20_+_34556528
|
1.695
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr7_+_102023016
|
1.516
|
|
|
|
|
chrX_-_13835313
|
1.381
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr19_+_48758931
|
1.369
|
|
LOC100505812
|
uncharacterized LOC100505812
|
|
chr12_-_10021932
|
1.244
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr2_+_132480063
|
0.998
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr10_-_113933517
|
0.996
|
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr17_-_34328530
|
0.981
|
|
CCL14-CCL15
CCL15
|
CCL14-CCL15 readthrough
chemokine (C-C motif) ligand 15
|
|
chr5_+_140557370
|
0.960
|
NM_019120
|
PCDHB8
|
protocadherin beta 8
|
|
chr2_-_99279935
|
0.935
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr6_-_25874439
|
0.915
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr22_+_33196801
|
0.907
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr7_-_16844606
|
0.886
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr5_+_140729723
|
0.879
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr10_+_49609641
|
0.856
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr7_+_94297448
|
0.845
|
|
PEG10
|
paternally expressed 10
|
|
chr3_-_9920633
|
0.838
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr3_-_194072004
|
0.837
|
NM_001080513
|
CPN2
|
carboxypeptidase N, polypeptide 2
|
|
chr12_-_11422640
|
0.831
|
NM_006249
|
PRB3
|
proline-rich protein BstNI subfamily 3
|
|
chr17_-_59668540
|
0.831
|
NM_199290
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2
|
|
chr5_+_140593508
|
0.820
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr11_-_124190092
|
0.800
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr22_+_36044412
|
0.798
|
NM_030641
|
APOL6
|
apolipoprotein L, 6
|
|
chr6_+_28317687
|
0.795
|
NM_001242894
NM_001242895
NM_024493
|
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3
|
|
chr3_-_126327397
|
0.774
|
NM_001039783
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor
|
|
chr5_+_76435073
|
0.772
|
|
|
|
|
chr17_-_18266691
|
0.755
|
NM_004169
NM_148918
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble)
|
|
chr2_-_77749335
|
0.751
|
NM_001134745
NM_024993
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr2_-_200213802
|
0.738
|
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_46747095
|
0.728
|
NM_080653
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2
|
|
chr11_-_6790187
|
0.726
|
NM_001004490
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2
|
|
chr12_+_131438451
|
0.724
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr13_-_41768392
|
0.720
|
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr7_-_115670774
|
0.708
|
|
TFEC
|
transcription factor EC
|
|
chr22_+_36044475
|
0.705
|
|
APOL6
|
apolipoprotein L, 6
|
|
chrX_+_107288199
|
0.689
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr18_-_14132426
|
0.685
|
NM_145287
|
ZNF519
|
zinc finger protein 519
|
|
chr12_+_9142136
|
0.683
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr1_+_54359859
|
0.681
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr2_-_233877920
|
0.666
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr6_+_27107060
|
0.652
|
NM_003495
|
HIST2H4B
HIST1H4I
|
histone cluster 2, H4b
histone cluster 1, H4i
|
|
chr10_-_93669184
|
0.648
|
NM_152429
|
FGFBP3
|
fibroblast growth factor binding protein 3
|
|
chr12_-_63546589
|
0.621
|
NM_000706
|
AVPR1A
|
arginine vasopressin receptor 1A
|
|
chr2_-_233877901
|
0.615
|
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr7_-_115670811
|
0.614
|
|
TFEC
|
transcription factor EC
|
|
chr5_+_140602914
|
0.611
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr5_+_36608430
|
0.610
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr17_-_34328607
|
0.610
|
|
CCL14-CCL15
|
CCL14-CCL15 readthrough
|
|
chr5_+_76382458
|
0.594
|
|
LOC728723
|
uncharacterized LOC728723
|
|
chr19_+_20959109
|
0.570
|
|
|
|
|
chr7_-_102283237
|
0.565
|
NM_001114403
|
UPK3BL
|
uroplakin 3B-like
|
|
chr18_-_48346297
|
0.564
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr2_-_27333963
|
0.536
|
NM_001166241
|
CGREF1
|
cell growth regulator with EF-hand domain 1
|
|
chr6_-_39072412
|
0.534
|
|
C6orf64
|
chromosome 6 open reading frame 64
|
|
chr11_+_5653484
|
0.533
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr4_-_159094163
|
0.520
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_140782367
|
0.517
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr1_-_32385258
|
0.515
|
NM_001195100
NM_001195101
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr5_-_35048239
|
0.514
|
NM_031900
|
AGXT2
|
alanine--glyoxylate aminotransferase 2
|
|
chr4_-_38858407
|
0.514
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr3_+_44692500
|
0.512
|
|
ZNF35
|
zinc finger protein 35
|
|
chr1_+_236557643
|
0.512
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr1_-_146697155
|
0.507
|
NM_001144829
NM_001144830
NM_001461
|
FMO5
|
flavin containing monooxygenase 5
|
|
chr4_-_100212085
|
0.507
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr10_+_74870209
|
0.502
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr7_-_115670828
|
0.501
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr19_+_36486042
|
0.499
|
NM_001042631
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1
|
|
chr1_+_155853546
|
0.499
|
|
SYT11
|
synaptotagmin XI
|
|
chr8_+_38585703
|
0.497
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr7_+_99425635
|
0.495
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr5_+_140514787
|
0.493
|
NM_015669
|
PCDHB5
|
protocadherin beta 5
|
|
chr19_-_23433161
|
0.492
|
|
ZNF724P
|
zinc finger protein 724, pseudogene
|
|
chr4_-_164534656
|
0.489
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr1_-_39347258
|
0.484
|
NM_030772
|
GJA9-MYCBP
GJA9
|
GJA9-MYCBP readthrough
gap junction protein, alpha 9, 59kDa
|
|
chr10_+_91152321
|
0.484
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr4_+_53185
|
0.483
|
NM_182524
|
ZNF595
ZNF718
|
zinc finger protein 595
zinc finger protein 718
|
|
chr5_+_86676620
|
0.469
|
|
|
|
|
chr2_+_204571197
|
0.459
|
NM_001243077
NM_001243078
NM_006139
|
CD28
|
CD28 molecule
|
|
chr5_+_140174443
|
0.447
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr8_-_33457435
|
0.447
|
NM_024025
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr19_-_23941571
|
0.446
|
NM_138286
|
ZNF681
|
zinc finger protein 681
|
|
chr19_+_12203077
|
0.444
|
|
ZNF788
|
zinc finger family member 788
|
|
chr12_+_70759974
|
0.443
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr11_+_7618416
|
0.436
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr5_+_36606667
|
0.435
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr5_-_147162251
|
0.429
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr19_-_23869939
|
0.411
|
NM_138330
|
ZNF675
|
zinc finger protein 675
|
|
chr11_+_56143099
|
0.411
|
NM_001005204
NM_001013356
|
OR8U1
OR8U8
|
olfactory receptor, family 8, subfamily U, member 1
olfactory receptor, family 8, subfamily U, member 8
|
|
chr3_-_168864325
|
0.407
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr21_+_38455246
|
0.405
|
NM_003316
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr4_+_156587861
|
0.402
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr17_-_16557157
|
0.401
|
NM_020787
|
ZNF624
|
zinc finger protein 624
|
|
chr2_-_88285308
|
0.400
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr6_+_161123224
|
0.390
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr3_-_57326709
|
0.374
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr4_-_155511838
|
0.373
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr12_-_54653317
|
0.371
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr5_+_140235594
|
0.368
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr12_-_11139510
|
0.367
|
NM_176890
|
TAS2R50
|
taste receptor, type 2, member 50
|
|
chr5_-_138718941
|
0.365
|
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr12_-_21927614
|
0.364
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr13_-_49018802
|
0.362
|
NM_005767
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr19_+_44455379
|
0.357
|
NM_013359
|
ZNF221
|
zinc finger protein 221
|
|
chr5_+_140743755
|
0.355
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr5_+_121465214
|
0.354
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr4_-_159093523
|
0.353
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr15_-_73051650
|
0.352
|
|
|
|
|
chr18_-_51880942
|
0.348
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr1_+_10490158
|
0.347
|
NM_198544
NM_199006
NM_199294
|
APITD1-CORT
APITD1
|
APITD1-CORT readthrough
apoptosis-inducing, TAF9-like domain 1
|
|
chr12_-_54653369
|
0.342
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr6_+_27777713
|
0.342
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chr12_+_29376596
|
0.341
|
NM_018099
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr10_-_69425356
|
0.339
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr6_-_49712055
|
0.336
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr19_+_1270926
|
0.334
|
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr1_+_236958690
|
0.332
|
|
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase
|
|
chr3_-_170744458
|
0.319
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr7_-_115608366
|
0.318
|
NM_001244583
|
TFEC
|
transcription factor EC
|
|
chr16_+_22217369
|
0.316
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr9_-_37592465
|
0.309
|
|
TOMM5
|
translocase of outer mitochondrial membrane 5 homolog (yeast)
|
|
chr13_+_108922205
|
0.306
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr1_-_182573393
|
0.300
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr2_+_88047603
|
0.298
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr3_-_12586962
|
0.295
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chrX_-_2886285
|
0.294
|
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr12_+_4699243
|
0.293
|
NM_003845
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4
|
|
chr1_+_74701070
|
0.293
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chrX_+_102965836
|
0.291
|
NM_182541
|
TMEM31
|
transmembrane protein 31
|
|
chr4_-_21699317
|
0.291
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr19_-_51872256
|
0.291
|
NM_152353
|
CLDND2
|
claudin domain containing 2
|
|
chr1_-_67266930
|
0.287
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr9_+_111624545
|
0.287
|
NM_006687
|
ACTL7A
|
actin-like 7A
|
|
chr14_-_23285011
|
0.284
|
NM_001126105
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr14_+_62462540
|
0.284
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr5_-_114961702
|
0.284
|
NM_001164468
NM_001164469
NM_181836
|
TMED7-TICAM2
TMED7
|
TMED7-TICAM2 readthrough
transmembrane emp24 protein transport domain containing 7
|
|
chr19_-_20150251
|
0.283
|
NM_033196
|
ZNF682
|
zinc finger protein 682
|
|
chr11_+_72975569
|
0.281
|
NM_176796
NM_176797
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr2_+_167744996
|
0.281
|
NM_001079810
NM_001199143
NM_152381
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr3_+_119187782
|
0.276
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr11_-_115088628
|
0.275
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr5_-_147162264
|
0.275
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr4_-_46125929
|
0.273
|
NM_173536
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1
|
|
chr8_-_33457491
|
0.270
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr2_-_74382352
|
0.269
|
|
|
|
|
chr4_+_120056938
|
0.267
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr17_-_48945338
|
0.266
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr2_-_188419185
|
0.263
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr16_+_15596122
|
0.262
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr1_+_92632608
|
0.259
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr5_+_61874561
|
0.256
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr3_-_48659188
|
0.256
|
NM_001008269
|
TMEM89
|
transmembrane protein 89
|
|
chr19_+_20277991
|
0.256
|
NM_052852
|
ZNF486
|
zinc finger protein 486
|
|
chr10_-_91102430
|
0.256
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr9_-_4742042
|
0.256
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr4_-_159956332
|
0.255
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr4_-_186578122
|
0.254
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr14_-_75600111
|
0.247
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr7_+_64838791
|
0.246
|
|
ZNF92
|
zinc finger protein 92
|
|
chr16_-_72206348
|
0.246
|
NM_001160213
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr20_+_43030005
|
0.244
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr12_-_46121489
|
0.241
|
|
LOC400027
|
uncharacterized LOC400027
|
|
chr6_-_2751153
|
0.241
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr14_+_74083547
|
0.241
|
NM_001037162
|
ACOT6
|
acyl-CoA thioesterase 6
|
|
chr20_+_43029973
|
0.240
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr1_-_163113938
|
0.239
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_-_89591685
|
0.238
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr18_+_21572736
|
0.237
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr10_+_135160843
|
0.236
|
NM_001145201
NM_145202
|
PRAP1
|
proline-rich acidic protein 1
|
|
chr3_-_186262114
|
0.233
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|
|
chr12_-_81991959
|
0.227
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr19_+_48216600
|
0.227
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr6_+_131958441
|
0.225
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr19_-_5870546
|
0.223
|
NM_002034
|
FUT5
|
fucosyltransferase 5 (alpha (1,3) fucosyltransferase)
|
|
chr20_+_43029865
|
0.219
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr20_-_18477811
|
0.218
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr17_+_9729380
|
0.216
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr3_-_123339423
|
0.215
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr1_+_161185084
|
0.214
|
NM_004106
|
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide
|
|
chr4_+_56261763
|
0.212
|
|
|
|
|
chr7_+_143792140
|
0.211
|
NM_001004135
|
OR2A12
|
olfactory receptor, family 2, subfamily A, member 12
|
|
chrX_-_10535476
|
0.210
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr22_+_38082331
|
0.208
|
NM_024313
|
NOL12
|
nucleolar protein 12
|
|
chr8_+_97505895
|
0.208
|
|
SDC2
|
syndecan 2
|
|
chr10_-_5045967
|
0.207
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr22_-_43485333
|
0.204
|
NM_012263
|
TTLL1
|
tubulin tyrosine ligase-like family, member 1
|
|
chr2_+_183943286
|
0.203
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr4_+_88896868
|
0.203
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr5_+_54455945
|
0.202
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr15_+_78857827
|
0.201
|
NM_000745
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5
|
|
chr1_+_206808751
|
0.200
|
NM_001004023
NM_003582
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr19_+_21324826
|
0.199
|
NM_133473
|
ZNF431
|
zinc finger protein 431
|
|
chr19_-_21950322
|
0.196
|
NM_173531
|
ZNF100
|
zinc finger protein 100
|
|
chr20_+_43803539
|
0.196
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr19_+_37180301
|
0.195
|
NM_152603
|
ZNF567
|
zinc finger protein 567
|
|
chr1_-_190446701
|
0.192
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr4_-_186577858
|
0.191
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_81771844
|
0.189
|
|
LPHN2
|
latrophilin 2
|