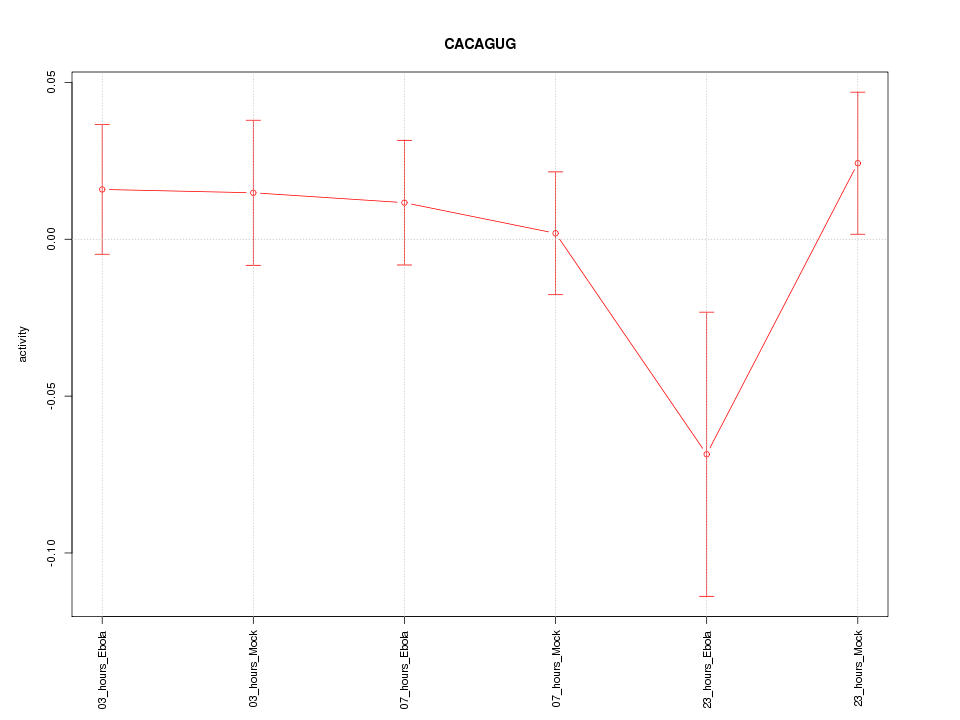
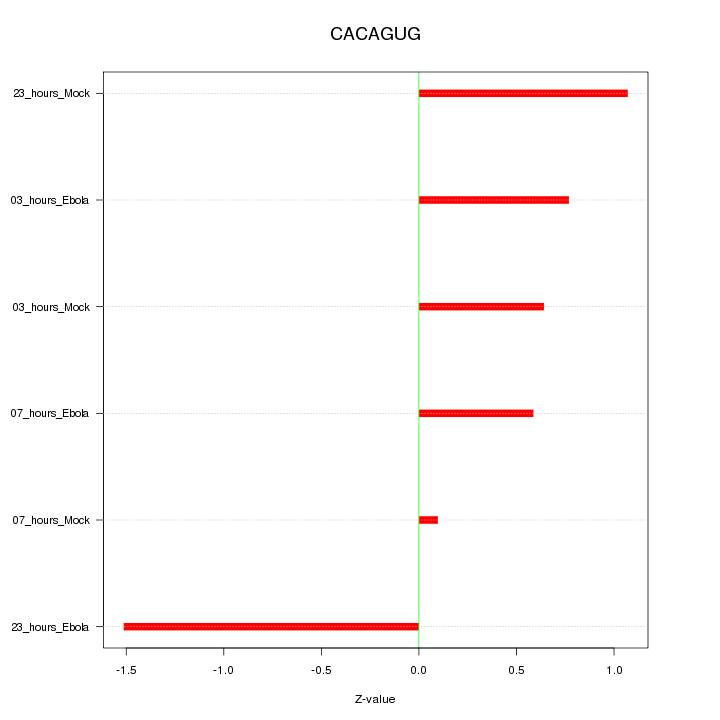
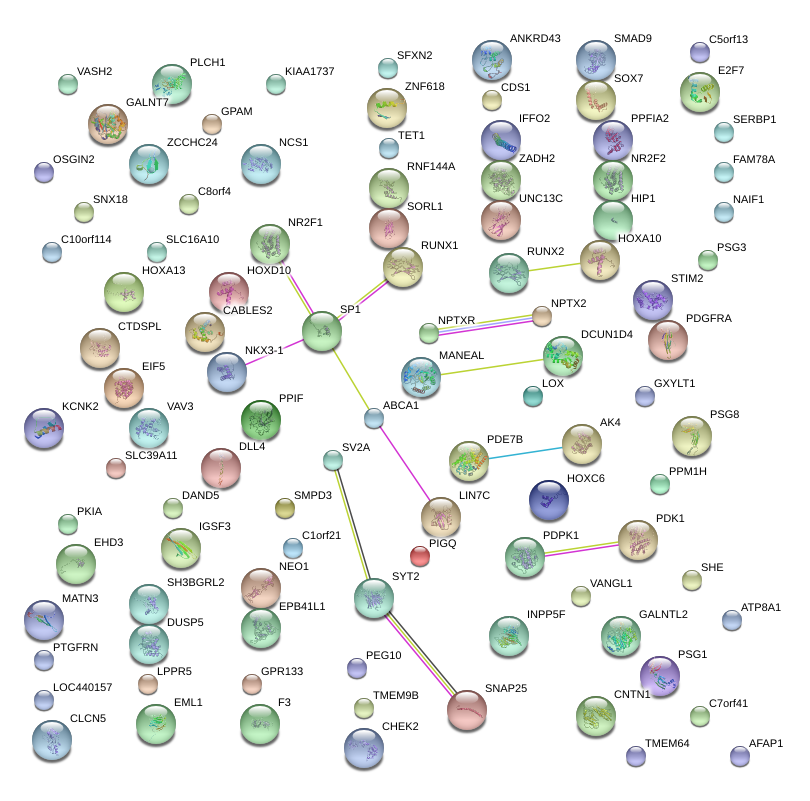
|
chr10_-_113943467
|
1.849
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr9_-_107690526
|
1.638
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr9_+_116638544
|
1.636
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr15_+_41221530
|
1.600
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr18_-_72921280
|
1.565
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr20_+_10199455
|
1.504
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr5_-_111093251
|
1.501
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_213123853
|
1.465
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr5_-_111093792
|
1.432
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_+_104474297
|
1.427
|
NM_178858
|
SFXN2
|
sideroflexin 2
|
|
chr5_-_111092918
|
1.377
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093030
|
1.371
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111091947
|
1.273
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_184356128
|
1.266
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr11_+_121322848
|
1.259
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr5_-_111312522
|
1.225
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_+_79428335
|
1.182
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr1_+_117452359
|
1.139
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr15_+_54305100
|
1.135
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr4_+_85504056
|
1.082
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr7_+_30174343
|
1.078
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr1_+_215256559
|
1.064
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr7_-_27239724
|
1.037
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr7_-_27213873
|
1.007
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr1_-_154474524
|
0.972
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr21_-_36421586
|
0.962
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_+_80340971
|
0.937
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr1_-_202679415
|
0.937
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr1_-_99470177
|
0.919
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr14_+_100259445
|
0.912
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr13_-_37494370
|
0.909
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr10_+_70320044
|
0.864
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr2_+_7057471
|
0.851
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr10_-_81205091
|
0.797
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr7_+_94285676
|
0.785
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr12_-_77459198
|
0.782
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr3_-_155394104
|
0.765
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr12_-_82152269
|
0.764
|
NM_001220475
NM_001220476
NM_001220473
NM_001220474
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr1_+_65613211
|
0.756
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr12_+_131438451
|
0.754
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr8_-_10587942
|
0.739
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr1_-_95007139
|
0.737
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr8_+_40010986
|
0.725
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr7_+_94285620
|
0.722
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr1_-_149889362
|
0.721
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr15_+_73344824
|
0.697
|
NM_001172623
NM_001172624
NM_002499
|
NEO1
|
neogenin 1
|
|
chr1_-_202612580
|
0.695
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr9_+_132934685
|
0.680
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr1_+_65613771
|
0.668
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr9_-_134151905
|
0.667
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr12_+_53773924
|
0.667
|
NM_001251825
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chr1_-_117210301
|
0.655
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr1_-_19282773
|
0.655
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr3_+_37903643
|
0.644
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr14_+_103801160
|
0.643
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr4_+_26862312
|
0.643
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr11_-_27528298
|
0.637
|
NM_018362
|
LIN7C
|
lin-7 homolog C (C. elegans)
|
|
chr1_+_116185249
|
0.633
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr10_-_21786205
|
0.617
|
NM_001010911
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr4_+_55095263
|
0.613
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr1_+_38259458
|
0.611
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr4_-_42658883
|
0.611
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr17_-_71088832
|
0.610
|
NM_001159770
NM_139177
|
SLC39A11
|
solute carrier family 39 (metal ion transporter), member 11
|
|
chr4_+_174089889
|
0.610
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr15_+_49170264
|
0.606
|
NM_014335
|
EID1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr5_+_132149022
|
0.606
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr22_-_39240016
|
0.600
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr3_-_155421996
|
0.599
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr12_-_42538432
|
0.596
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr4_-_7941092
|
0.594
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr5_-_121414011
|
0.587
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr11_-_8985955
|
0.582
|
NM_020644
|
TMEM9B
|
TMEM9 domain family, member B
|
|
chr8_-_91657898
|
0.581
|
NM_001008495
NM_001146273
|
TMEM64
|
transmembrane protein 64
|
|
chr2_+_31456879
|
0.577
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr1_+_116184573
|
0.576
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr4_+_52709165
|
0.575
|
NM_001040402
NM_015115
|
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr20_-_60982334
|
0.567
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr2_+_173420776
|
0.565
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr14_+_103800492
|
0.562
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr1_-_108231122
|
0.556
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr9_-_130829598
|
0.556
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr8_-_23540401
|
0.555
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chrX_+_49832214
|
0.555
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr20_+_34742656
|
0.551
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr6_+_111408780
|
0.550
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr12_-_63328663
|
0.546
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr14_+_77564570
|
0.539
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr15_+_96873845
|
0.539
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr10_+_112257624
|
0.536
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr3_+_119187782
|
0.534
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr6_+_136172802
|
0.520
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr12_+_54422193
|
0.517
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr5_+_53813494
|
0.515
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chr7_+_98246595
|
0.511
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr10_+_81107219
|
0.506
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr15_+_96869156
|
0.506
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_67896098
|
0.504
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr7_-_75368276
|
0.500
|
NM_001243198
NM_005338
|
HIP1
|
huntingtin interacting protein 1
|
|
chr16_-_68482373
|
0.497
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr1_-_108507474
|
0.496
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr2_-_20212448
|
0.495
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr5_+_176560056
|
0.494
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr2_+_157291951
|
0.491
|
NM_001083112
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
|
|
chr3_+_113465860
|
0.489
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr12_-_47219734
|
0.488
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr5_-_111754738
|
0.481
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chr13_-_80915041
|
0.481
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr3_+_67048726
|
0.481
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr15_+_29213839
|
0.479
|
NM_001130414
NM_005503
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2
|
|
chrX_-_3631654
|
0.476
|
NM_005044
|
PRKX
|
protein kinase, X-linked
|
|
chr9_+_101867401
|
0.472
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr15_+_96876568
|
0.471
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr4_-_146859568
|
0.467
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr17_-_53499055
|
0.465
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr15_-_63674074
|
0.465
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr7_+_73498106
|
0.465
|
NM_002314
|
LIMK1
|
LIM domain kinase 1
|
|
chr2_+_202899309
|
0.463
|
NM_003507
|
FZD7
|
frizzled family receptor 7
|
|
chr17_+_28705922
|
0.452
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr6_-_42419782
|
0.450
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr6_+_114178516
|
0.449
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr12_-_81991959
|
0.447
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr8_+_132916339
|
0.447
|
NM_015137
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr2_+_157292826
|
0.445
|
NM_000408
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
|
|
chr12_+_53774422
|
0.441
|
NM_003109
|
SP1
|
Sp1 transcription factor
|
|
chr11_+_19372270
|
0.441
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr7_-_23053732
|
0.441
|
NM_032581
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr12_-_93835986
|
0.439
|
NM_003348
|
UBE2N
|
ubiquitin-conjugating enzyme E2N
|
|
chr2_-_25141855
|
0.436
|
NM_004036
|
ADCY3
|
adenylate cyclase 3
|
|
chr9_+_118916069
|
0.427
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr12_-_109747024
|
0.424
|
NM_213596
|
FOXN4
|
forkhead box N4
|
|
chr5_-_121412805
|
0.424
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr2_-_71454200
|
0.422
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr4_-_53525316
|
0.422
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr11_+_12695851
|
0.420
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr1_+_145438437
|
0.419
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr20_+_62795826
|
0.417
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr3_+_122920773
|
0.417
|
NM_012430
|
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae)
|
|
chr5_+_176560627
|
0.412
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr9_-_136242969
|
0.411
|
NM_033161
|
SURF4
|
surfeit 4
|
|
chr3_-_129612371
|
0.408
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr1_-_41131323
|
0.407
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr19_+_38397841
|
0.406
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr3_-_142682177
|
0.404
|
NM_198504
|
PAQR9
|
progestin and adipoQ receptor family member IX
|
|
chr7_+_116312412
|
0.401
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr2_-_191399449
|
0.399
|
NM_001142645
|
TMEM194B
|
transmembrane protein 194B
|
|
chr1_+_38261079
|
0.399
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr6_-_107435635
|
0.398
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chrX_+_150151757
|
0.396
|
NM_005342
|
HMGB3
|
high mobility group box 3
|
|
chr12_-_81763127
|
0.396
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr4_+_113970784
|
0.395
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr12_-_105478218
|
0.392
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chr7_+_73507485
|
0.391
|
NM_001204426
|
LIMK1
|
LIM domain kinase 1
|
|
chr3_-_113415313
|
0.391
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr17_-_56084656
|
0.386
|
NM_001078166
NM_006924
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr7_-_92219661
|
0.385
|
NM_001040057
NM_152789
|
FAM133B
|
family with sequence similarity 133, member B
|
|
chr17_-_40761404
|
0.383
|
NM_178126
|
FAM134C
|
family with sequence similarity 134, member C
|
|
chr1_+_215178884
|
0.380
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr18_+_55102777
|
0.380
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr7_-_139477437
|
0.379
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr4_+_6784453
|
0.378
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr6_+_170102236
|
0.378
|
NM_001029863
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr1_-_86043932
|
0.377
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_184724021
|
0.377
|
NM_025191
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr5_+_65017993
|
0.373
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr10_+_104503724
|
0.372
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr7_+_100136784
|
0.372
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr12_-_123451031
|
0.369
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr1_-_21605984
|
0.369
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr1_-_21616648
|
0.365
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr4_+_113739203
|
0.365
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_+_14444083
|
0.364
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr12_-_54653317
|
0.364
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr12_-_54653369
|
0.364
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr6_-_33160199
|
0.363
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr11_+_118401755
|
0.362
|
NM_001144034
NM_001144035
NM_001144036
NM_001144037
NM_001144038
NM_032780
|
TMEM25
|
transmembrane protein 25
|
|
chr16_-_1429612
|
0.362
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr1_+_24286300
|
0.360
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr3_-_49203769
|
0.358
|
NM_022903
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr15_+_96875793
|
0.358
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr19_+_51815095
|
0.357
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr22_+_46546498
|
0.356
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr20_+_43514340
|
0.350
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr16_-_53737713
|
0.349
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr20_+_58515408
|
0.348
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr4_-_53522756
|
0.348
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr3_+_137906080
|
0.347
|
NM_014154
NM_015396
NM_213654
|
ARMC8
|
armadillo repeat containing 8
|
|
chr8_-_41522724
|
0.346
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr20_-_48532067
|
0.345
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr12_+_54410641
|
0.343
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr8_-_41655139
|
0.343
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr5_+_14664764
|
0.343
|
NM_138348
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr9_+_129089092
|
0.343
|
NM_001011703
NM_033446
|
FAM125B
|
family with sequence similarity 125, member B
|
|
chr14_+_51706885
|
0.342
|
NM_030755
|
TMX1
|
thioredoxin-related transmembrane protein 1
|
|
chr11_+_77300679
|
0.342
|
NM_173039
|
AQP11
|
aquaporin 11
|
|
chr3_-_52931595
|
0.341
|
NM_001198974
NM_198563
|
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough
transmembrane protein 110
|
|
chr1_+_54519259
|
0.338
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr13_-_49018802
|
0.334
|
NM_005767
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr8_-_127570710
|
0.334
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chrX_+_17393537
|
0.334
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr1_-_85930733
|
0.334
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|