|
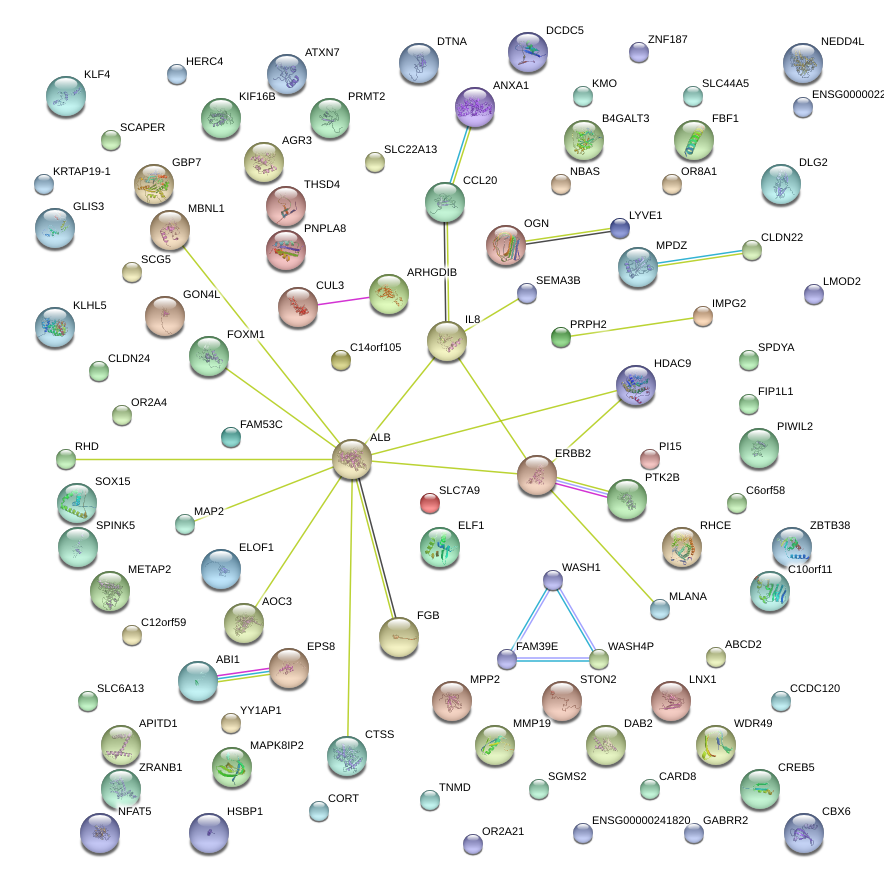
chr4_+_74606222
|
1.345
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr7_+_18535817
|
1.067
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr17_-_7493389
|
1.038
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr7_+_28452143
|
0.955
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_18535351
|
0.924
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr2_+_210444364
|
0.865
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr9_+_5890801
|
0.816
|
NM_005511
|
MLANA
|
melan-A
|
|
chr12_-_371994
|
0.748
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr5_+_137673223
|
0.746
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr1_-_89641722
|
0.733
|
NM_207398
|
GBP7
|
guanylate binding protein 7
|
|
chr9_+_75766777
|
0.613
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr2_+_29038862
|
0.563
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr4_+_54292035
|
0.533
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr15_+_32933869
|
0.517
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr9_-_110252045
|
0.507
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr9_-_29738
|
0.499
|
NM_182905
|
WASH1
|
WAS protein family homolog 1
|
|
chr9_+_75766794
|
0.473
|
|
ANXA1
|
annexin A1
|
|
chr22_+_51041561
|
0.466
|
NM_016431
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr17_+_41003184
|
0.446
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr2_+_228678556
|
0.423
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr15_+_71839540
|
0.423
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr7_+_123295860
|
0.412
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr10_+_126630691
|
0.398
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr1_-_150738292
|
0.375
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr1_-_150738255
|
0.352
|
|
CTSS
|
cathepsin S
|
|
chr16_+_69680959
|
0.349
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr7_-_16921596
|
0.349
|
NM_176813
|
AGR3
|
anterior gradient 3 homolog (Xenopus laevis)
|
|
chr19_-_33360641
|
0.323
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chr11_+_124439892
|
0.321
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr2_-_225370687
|
0.319
|
|
CUL3
|
cullin 3
|
|
chr7_+_28338939
|
0.318
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_+_127898318
|
0.317
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chrX_+_99839789
|
0.312
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr15_-_77154284
|
0.303
|
NM_001145923
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr12_-_111893857
|
0.301
|
|
|
|
|
chr1_-_150738232
|
0.298
|
|
CTSS
|
cathepsin S
|
|
chr4_+_155484131
|
0.294
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr12_-_15114495
|
0.290
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr18_+_55888838
|
0.289
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_-_13250313
|
0.284
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chr12_-_56236614
|
0.275
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr3_+_38307297
|
0.275
|
NM_004256
|
SLC22A13
|
solute carrier family 22 (organic anion transporter), member 13
|
|
chr4_-_184241926
|
0.271
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr7_+_28475233
|
0.270
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr22_-_39268230
|
0.268
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr12_-_40013842
|
0.266
|
NM_005164
|
ABCD2
|
ATP-binding cassette, sub-family D (ALD), member 2
|
|
chr9_-_110251308
|
0.265
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr4_+_108814419
|
0.264
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr13_-_41556360
|
0.261
|
NM_001145353
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr1_+_241695433
|
0.259
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr8_+_75736771
|
0.258
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr4_+_39063851
|
0.257
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr11_-_83984230
|
0.254
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr6_-_90024936
|
0.253
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr13_-_41556188
|
0.250
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr12_+_10331556
|
0.248
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr8_+_22210311
|
0.248
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr7_-_108142934
|
0.246
|
|
PNPLA8
|
patatin-like phospholipase domain containing 8
|
|
chr1_+_25598883
|
0.245
|
NM_001127691
NM_016124
|
RHD
|
Rh blood group, D antigen
|
|
chr10_-_27057776
|
0.238
|
|
ABI1
|
abl-interactor 1
|
|
chr12_-_15815073
|
0.237
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr3_+_63884074
|
0.234
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr10_+_77542518
|
0.231
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr11_-_31014232
|
0.230
|
NM_020869
|
DCDC5
|
doublecortin domain containing 5
|
|
chr18_+_32335939
|
0.229
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr19_-_48759141
|
0.226
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr21_-_31852631
|
0.224
|
NM_181607
|
KRTAP19-1
|
keratin associated protein 19-1
|
|
chr4_+_74286846
|
0.223
|
|
ALB
|
albumin
|
|
chr11_-_10590078
|
0.218
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr14_-_57960432
|
0.216
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr1_-_155774769
|
0.213
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr1_+_10509775
|
0.212
|
NM_001302
|
CORT
|
cortistatin
|
|
chrX_+_48916515
|
0.212
|
NM_001163322
NM_033626
NM_001163321
NM_001163323
|
CCDC120
|
coiled-coil domain containing 120
|
|
chr11_-_84634437
|
0.211
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr8_+_27179918
|
0.209
|
NM_004103
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr6_+_28234787
|
0.208
|
NM_001023560
NM_001111039
NM_152736
|
ZNF187
|
zinc finger protein 187
|
|
chr7_-_143956720
|
0.204
|
NM_001005328
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7
|
|
chr15_-_77176191
|
0.204
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr9_-_4152182
|
0.202
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr1_-_76076762
|
0.201
|
NM_001130058
NM_152697
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr1_-_161147757
|
0.199
|
NM_001199873
NM_001199874
NM_003779
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr3_+_141043054
|
0.198
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr3_-_167371288
|
0.196
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr2_-_15701445
|
0.195
|
NM_015909
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr18_+_32398225
|
0.191
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr9_-_95166827
|
0.188
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr3_-_101039403
|
0.185
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr5_-_39394423
|
0.183
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr5_+_147443534
|
0.182
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr21_+_48055077
|
0.181
|
|
PRMT2
|
protein arginine methyltransferase 2
|
|
chr17_+_37856224
|
0.179
|
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr3_+_151985828
|
0.179
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr14_-_81864726
|
0.178
|
NM_033104
|
STON2
|
stonin 2
|
|
chr10_-_69752385
|
0.178
|
|
HERC4
|
hect domain and RLD 4
|
|
chr3_+_50306578
|
0.178
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr9_+_130026755
|
0.177
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr10_+_63808968
|
0.176
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr17_+_76000248
|
0.176
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr6_-_149969849
|
0.175
|
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1
|
|
chr8_+_76452202
|
0.174
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr10_-_97200895
|
0.172
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr9_+_96846804
|
0.170
|
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr6_+_7402879
|
0.170
|
NM_153005
|
RIOK1
|
RIO kinase 1 (yeast)
|
|
chr15_+_83477972
|
0.166
|
NM_001080435
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chr10_-_74283693
|
0.166
|
NM_001195519
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr13_+_50025687
|
0.166
|
NM_001160308
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr11_+_112047088
|
0.165
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr17_+_72429835
|
0.164
|
NM_018653
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr7_-_100076754
|
0.163
|
NM_030935
|
TSC22D4
|
TSC22 domain family, member 4
|
|
chr10_-_65028825
|
0.162
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr21_+_35736322
|
0.161
|
NM_172201
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2
|
|
chr17_+_37856174
|
0.159
|
|
|
|
|
chr12_-_58165877
|
0.159
|
NM_005371
NM_023033
|
METTL1
|
methyltransferase like 1
|
|
chr12_+_28410132
|
0.158
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr1_+_78470593
|
0.157
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr4_+_62362838
|
0.157
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chrX_+_65384078
|
0.157
|
NM_001130860
NM_014799
|
HEPH
|
hephaestin
|
|
chr4_-_170077807
|
0.155
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr5_-_160279045
|
0.155
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr6_+_20534686
|
0.152
|
NM_017774
|
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1
|
|
chr3_-_49967194
|
0.151
|
NM_001142501
NM_032355
|
MON1A
|
MON1 homolog A (yeast)
|
|
chrM_+_10123
|
0.151
|
|
|
|
|
chr7_+_129269916
|
0.150
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr18_-_24445652
|
0.149
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr20_+_42984088
|
0.148
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr3_-_167371701
|
0.148
|
|
WDR49
|
WD repeat domain 49
|
|
chr3_-_151176496
|
0.148
|
NM_178822
|
IGSF10
|
immunoglobulin superfamily, member 10
|
|
chr2_+_143635225
|
0.146
|
|
KYNU
|
kynureninase
|
|
chr5_-_98262217
|
0.144
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr3_-_49726081
|
0.139
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr17_+_37856253
|
0.138
|
NM_004448
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr3_-_114790221
|
0.137
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_+_37892786
|
0.137
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr10_-_52645430
|
0.137
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chrM_+_5245
|
0.137
|
|
|
|
|
chr6_-_139613114
|
0.137
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr10_+_70482210
|
0.133
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr15_+_72406598
|
0.133
|
NM_001166340
|
SENP8
|
SUMO/sentrin specific peptidase family member 8
|
|
chr19_-_10426673
|
0.132
|
NM_001031734
|
FDX1L
|
ferredoxin 1-like
|
|
chr14_-_37641864
|
0.132
|
NM_001171170
NM_030631
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21
|
|
chr2_+_108994366
|
0.132
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr4_+_109543670
|
0.132
|
|
RPL34
|
ribosomal protein L34
|
|
chr2_-_100722328
|
0.131
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chrX_-_102774416
|
0.131
|
NM_080879
|
RAB40A
|
RAB40A, member RAS oncogene family
|
|
chr7_-_111428944
|
0.127
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr6_-_46293215
|
0.125
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_-_59147727
|
0.125
|
|
|
|
|
chr3_-_87325611
|
0.125
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr13_+_78315294
|
0.124
|
NM_001242869
NM_001242870
NM_001242871
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr21_+_38792600
|
0.124
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr13_+_97928414
|
0.122
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr3_+_102153858
|
0.122
|
NM_175056
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr2_-_216241321
|
0.122
|
|
FN1
|
fibronectin 1
|
|
chr4_+_72897520
|
0.121
|
NM_004885
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chr1_-_231560789
|
0.121
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr2_+_143635194
|
0.120
|
NM_001032998
NM_001199241
NM_003937
|
KYNU
|
kynureninase
|
|
chrX_-_24045302
|
0.120
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr17_+_73028670
|
0.119
|
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr5_+_145316120
|
0.119
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr6_+_24357130
|
0.118
|
NM_181337
|
KAAG1
|
kidney associated antigen 1
|
|
chr2_+_74120132
|
0.118
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr1_+_178310605
|
0.117
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr11_-_16497917
|
0.117
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr10_-_62493077
|
0.116
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr5_+_145317297
|
0.115
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr16_+_4845265
|
0.113
|
NM_001253793
NM_001253794
|
LOC440335
|
uncharacterized LOC440335
|
|
chr6_-_33754712
|
0.112
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr14_+_76452053
|
0.111
|
NM_001102564
NM_052873
|
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas)
|
|
chr11_-_84028379
|
0.110
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr14_-_20801427
|
0.110
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr3_+_151488214
|
0.109
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr15_-_41364176
|
0.108
|
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr5_-_37184945
|
0.107
|
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr13_+_97794050
|
0.103
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_-_132200476
|
0.103
|
NM_005260
|
GDF9
|
growth differentiation factor 9
|
|
chr9_-_4666665
|
0.103
|
NM_001039395
|
C9orf68
|
chromosome 9 open reading frame 68
|
|
chr9_+_96846710
|
0.102
|
NM_001253829
NM_152422
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr16_-_3422282
|
0.102
|
NM_001190476
|
MTRNR2L4
|
MT-RNR2-like 4
|
|
chr13_-_41593405
|
0.101
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr18_+_32173281
|
0.099
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_+_71587674
|
0.099
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr12_-_10542616
|
0.099
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chrX_+_119737743
|
0.097
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_+_151129104
|
0.097
|
NM_001204848
NM_024575
|
TNFAIP8L2-SCNM1
TNFAIP8L2
|
TNFAIP8L2-SCNM1 readthrough
tumor necrosis factor, alpha-induced protein 8-like 2
|
|
chr20_-_30539725
|
0.097
|
|
PDRG1
|
p53 and DNA-damage regulated 1
|
|
chr4_-_15939949
|
0.097
|
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr5_+_141346401
|
0.097
|
NM_001201365
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr19_-_48752921
|
0.095
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr6_-_133084585
|
0.095
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr9_-_104145800
|
0.095
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr5_+_149887673
|
0.095
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr4_+_187187342
|
0.095
|
|
F11
|
coagulation factor XI
|
|
chr1_-_42776760
|
0.094
|
|
|
|
|
chr3_-_187452669
|
0.093
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_-_48115846
|
0.093
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr1_+_192544856
|
0.093
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr11_-_3803310
|
0.093
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr3_+_183353355
|
0.092
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr7_+_112063131
|
0.092
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr15_+_45028559
|
0.092
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|