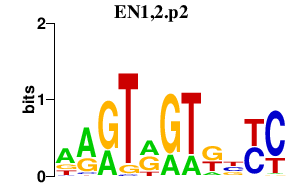
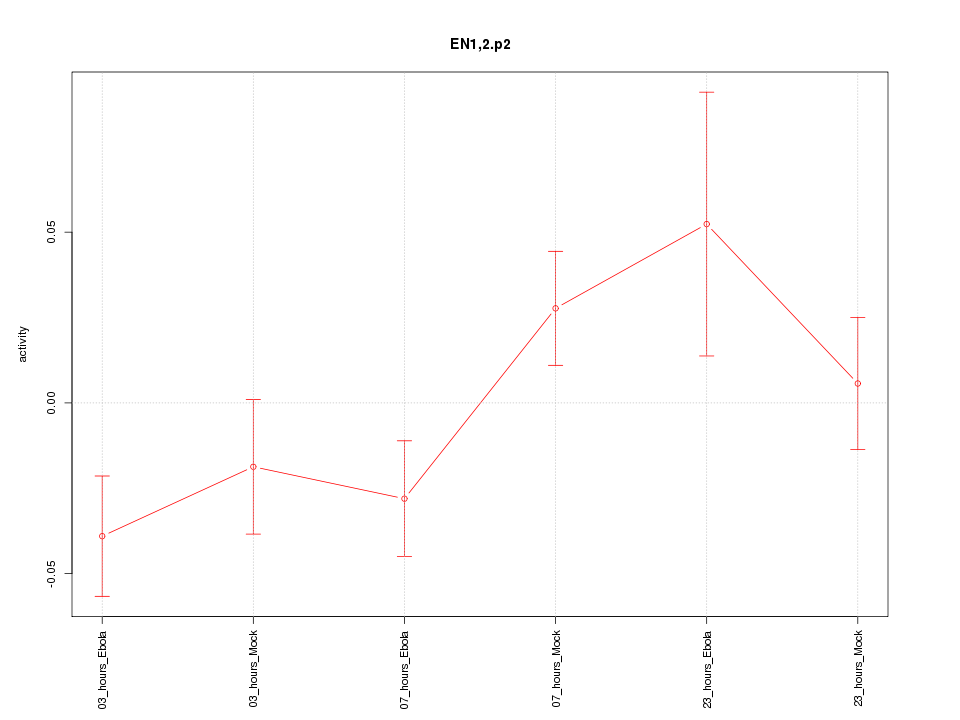
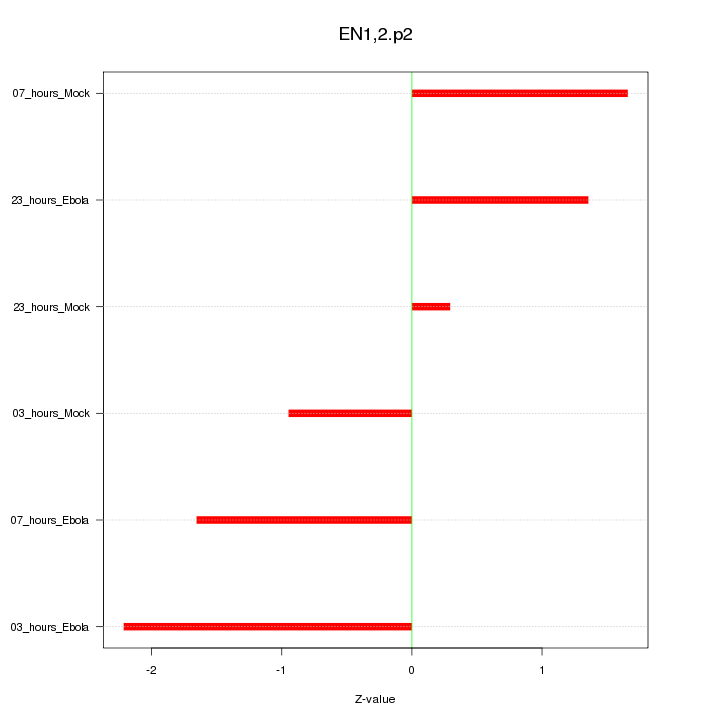
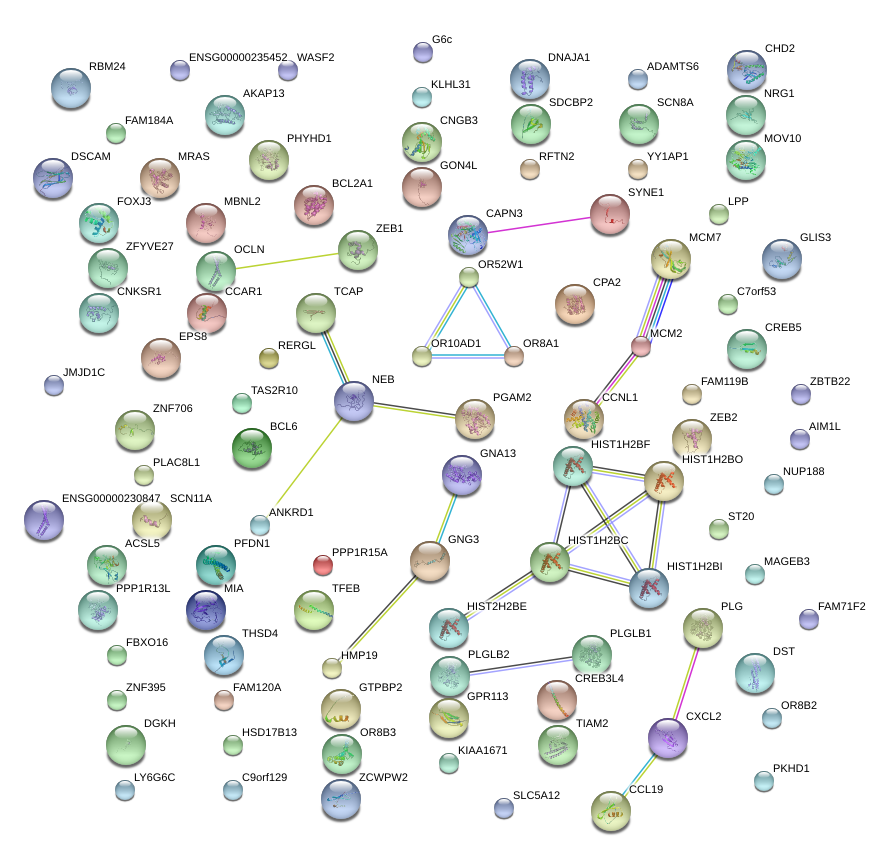
|
chr6_+_17282931
|
3.407
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_-_87248942
|
2.064
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chrM_+_10123
|
1.914
|
|
|
|
|
chr2_-_152590983
|
1.626
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr2_-_145277879
|
1.365
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr17_-_63009302
|
1.295
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr11_+_62475113
|
1.259
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr9_+_33026482
|
1.202
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr6_-_41703938
|
1.155
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr7_+_28452143
|
1.103
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_-_43595079
|
1.093
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr6_-_43595046
|
1.073
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr17_+_37821598
|
1.016
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr19_+_41281281
|
1.012
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr8_-_87755880
|
1.004
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr15_+_71433787
|
0.905
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr13_+_42712177
|
0.874
|
NM_001204505
NM_001204506
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr17_+_7384720
|
0.838
|
NM_001102614
|
SLC35G6
|
solute carrier family 35, member G6
|
|
chr19_+_49376488
|
0.809
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr8_+_32579350
|
0.806
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr13_+_97874540
|
0.765
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr10_-_92680784
|
0.751
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_+_74920329
|
0.742
|
|
|
|
|
chr7_+_28530747
|
0.740
|
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_-_187452669
|
0.738
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr6_-_152702450
|
0.730
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr11_+_6220375
|
0.715
|
NM_001005178
|
OR52W1
|
olfactory receptor, family 52, subfamily W, member 1
|
|
chr11_-_124253238
|
0.700
|
NM_001005468
|
OR8B2
|
olfactory receptor, family 8, subfamily B, member 2
|
|
chr10_+_114154522
|
0.662
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr6_-_51952368
|
0.654
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr6_-_30652434
|
0.646
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr3_-_156878481
|
0.636
|
NM_020307
|
CCNL1
|
cyclin L1
|
|
chr9_-_34691135
|
0.620
|
NM_006274
|
CCL19
|
chemokine (C-C motif) ligand 19
|
|
chr15_-_80263460
|
0.615
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr1_-_27732069
|
0.614
|
|
WASF2
|
WAS protein family, member 2
|
|
chr20_-_1309782
|
0.612
|
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr5_-_145483877
|
0.609
|
NM_001029869
|
PLAC8L1
|
PLAC8-like 1
|
|
chr9_+_131684561
|
0.591
|
NM_001100877
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr7_+_112120907
|
0.534
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr3_-_38992051
|
0.524
|
NM_014139
|
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit
|
|
chr6_-_53530505
|
0.511
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr3_+_28431986
|
0.507
|
NM_001040432
|
ZCWPW2
|
zinc finger, CW type with PWWP domain 2
|
|
chr1_+_16259046
|
0.493
|
|
|
|
|
chr1_+_153940345
|
0.488
|
NM_130898
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4
|
|
chr20_-_30288094
|
0.485
|
|
|
|
|
chr6_+_155411422
|
0.473
|
NM_012454
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr12_-_10978867
|
0.470
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chr12_-_15865847
|
0.467
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr6_+_26273143
|
0.466
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr15_+_42696957
|
0.459
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr5_-_64777703
|
0.456
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chrX_+_30248552
|
0.455
|
NM_002365
|
MAGEB3
|
melanoma antigen family B, 3
|
|
chr2_-_198540583
|
0.455
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr10_-_65225605
|
0.454
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr19_-_45908288
|
0.452
|
NM_006663
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr6_-_56507585
|
0.448
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr5_+_173472692
|
0.446
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr12_-_48597074
|
0.444
|
NM_001004134
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1
|
|
chr12_-_18243052
|
0.439
|
NM_024730
|
RERGL
|
RERG/RAS-like
|
|
chr10_+_114133838
|
0.438
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr5_+_68800003
|
0.432
|
NM_001205255
|
OCLN
|
occludin
|
|
chr15_+_93443559
|
0.430
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr1_-_155774769
|
0.424
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr10_+_114133775
|
0.421
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr7_-_44105162
|
0.420
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr3_+_187870808
|
0.419
|
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr7_+_129906702
|
0.419
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr15_+_93443418
|
0.407
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr3_+_138066489
|
0.406
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr11_+_65266642
|
0.403
|
|
|
|
|
chr12_-_57674251
|
0.397
|
|
|
|
|
chr6_-_119470336
|
0.395
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr4_-_88243994
|
0.395
|
NM_001136230
NM_178135
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13
|
|
chr1_+_113236679
|
0.394
|
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse)
|
|
chr1_-_26680620
|
0.382
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chr2_-_26541916
|
0.380
|
NM_001145168
NM_001145169
|
GPR113
|
G protein-coupled receptor 113
|
|
chr9_-_4152182
|
0.378
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr11_-_26743545
|
0.375
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr5_-_139682657
|
0.373
|
|
|
|
|
chr9_-_96108695
|
0.373
|
NM_001098808
|
C9orf129
|
chromosome 9 open reading frame 129
|
|
chr10_+_70482210
|
0.373
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr15_+_86220267
|
0.370
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr10_+_99498233
|
0.368
|
NM_001002261
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr7_+_128312345
|
0.361
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr22_+_25591072
|
0.359
|
|
KIAA1671
|
KIAA1671
|
|
chr6_-_31689455
|
0.355
|
NM_025261
|
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr8_+_11188494
|
0.354
|
NM_054028
|
SLC35G5
|
solute carrier family 35, member G5
|
|
chr19_+_41281081
|
0.354
|
NM_001202553
|
MIA
|
melanoma inhibitory activity
|
|
chr11_+_124439892
|
0.349
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr1_-_42800902
|
0.347
|
NM_014947
|
FOXJ3
|
forkhead box J3
|
|
chr5_-_139682674
|
0.342
|
|
PFDN1
|
prefoldin subunit 1
|
|
chr1_-_42801529
|
0.341
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chr14_-_77889231
|
0.341
|
NM_001113475
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1
|
|
chr1_-_150587879
|
0.340
|
|
|
|
|
chr8_-_28347733
|
0.339
|
NM_172366
|
FBXO16
ZNF395
|
F-box protein 16
zinc finger protein 395
|
|
chr2_+_88047603
|
0.333
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr4_-_74964996
|
0.332
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr12_+_52056547
|
0.328
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr1_+_26503973
|
0.327
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr4_-_54457723
|
0.326
|
NM_001126328
|
LNX1
|
ligand of numb-protein X 1
|
|
chr13_+_97874704
|
0.326
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_-_42800883
|
0.318
|
NM_001198852
|
FOXJ3
|
forkhead box J3
|
|
chr1_+_33231260
|
0.316
|
|
KIAA1522
|
KIAA1522
|
|
chr18_+_32398225
|
0.311
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr17_-_40828441
|
0.310
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr2_-_145277568
|
0.307
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_+_69488154
|
0.306
|
NM_004312
|
ARR3
|
arrestin 3, retinal (X-arrestin)
|
|
chr1_-_150780705
|
0.303
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr10_+_104179570
|
0.303
|
NM_024326
|
FBXL15
|
F-box and leucine-rich repeat protein 15
|
|
chr1_+_67632168
|
0.301
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr4_+_71062241
|
0.297
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr12_-_49999259
|
0.295
|
NM_032130
|
FAM186B
|
family with sequence similarity 186, member B
|
|
chr1_+_212797788
|
0.293
|
NM_153606
|
FAM71A
|
family with sequence similarity 71, member A
|
|
chr1_+_162351519
|
0.292
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr19_-_58067724
|
0.291
|
NM_001039654
|
ZNF550
|
zinc finger protein 550
|
|
chr15_-_77176191
|
0.291
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr3_+_69812620
|
0.290
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_33231227
|
0.288
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr12_-_57630867
|
0.285
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr16_+_31085751
|
0.282
|
|
ZNF646
|
zinc finger protein 646
|
|
chr7_+_28338939
|
0.281
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_-_141673572
|
0.280
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr1_-_42800623
|
0.279
|
NM_001198851
|
FOXJ3
|
forkhead box J3
|
|
chr6_-_31939658
|
0.278
|
|
DOM3Z
|
dom-3 homolog Z (C. elegans)
|
|
chr16_+_31085722
|
0.277
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr3_-_51996855
|
0.277
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr12_-_57914287
|
0.277
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr2_+_201170603
|
0.274
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr20_+_44036940
|
0.273
|
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr16_+_31494438
|
0.271
|
NM_003041
|
SLC5A2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2
|
|
chr11_-_6790187
|
0.270
|
NM_001004490
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2
|
|
chr15_-_45003513
|
0.270
|
|
|
|
|
chr11_+_47290893
|
0.267
|
NM_130470
|
MADD
|
MAP-kinase activating death domain
|
|
chr17_-_62777621
|
0.265
|
|
LOC146880
|
Rho GTPase activating protein 27 pseudogene
|
|
chr11_+_6128913
|
0.262
|
NM_001005181
|
OR56B4
|
olfactory receptor, family 56, subfamily B, member 4
|
|
chr14_-_61191027
|
0.261
|
|
SIX4
|
SIX homeobox 4
|
|
chr11_-_89956531
|
0.260
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr11_-_83984230
|
0.259
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_+_52813936
|
0.257
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr12_+_7033625
|
0.257
|
NM_001007026
|
ATN1
|
atrophin 1
|
|
chr12_-_22089627
|
0.255
|
NM_005691
NM_020297
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr6_-_109775423
|
0.254
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr4_+_103422479
|
0.254
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr10_+_13142055
|
0.253
|
NM_001008211
NM_001008212
NM_001008213
NM_021980
|
OPTN
|
optineurin
|
|
chr9_-_34662629
|
0.251
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr11_-_117100551
|
0.249
|
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr7_+_112092112
|
0.249
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr15_+_45021218
|
0.248
|
|
TRIM69
|
tripartite motif containing 69
|
|
chr20_+_54987167
|
0.248
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr7_+_107110501
|
0.248
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr13_+_73356179
|
0.247
|
NM_006346
|
PIBF1
|
progesterone immunomodulatory binding factor 1
|
|
chr5_+_137673223
|
0.245
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr1_-_145715561
|
0.243
|
NM_007053
|
CD160
|
CD160 molecule
|
|
chr5_-_139682680
|
0.243
|
NM_002622
|
PFDN1
|
prefoldin subunit 1
|
|
chr11_-_59633942
|
0.240
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr11_-_5863126
|
0.240
|
NM_001005167
|
OR52E6
|
olfactory receptor, family 52, subfamily E, member 6
|
|
chr6_-_41006927
|
0.239
|
NM_173561
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like
|
|
chr1_-_153919140
|
0.239
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr7_-_120498128
|
0.236
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr17_-_27467395
|
0.236
|
|
MYO18A
|
myosin XVIIIA
|
|
chr1_+_150039815
|
0.233
|
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae)
|
|
chr10_-_62149487
|
0.232
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr4_-_54457628
|
0.232
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr2_-_161056573
|
0.232
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr3_-_184095927
|
0.232
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr4_+_91156181
|
0.231
|
NM_207491
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr4_+_74281884
|
0.231
|
|
ALB
|
albumin
|
|
chr7_-_20256942
|
0.231
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr21_-_34883620
|
0.230
|
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr19_-_46295785
|
0.230
|
NM_004943
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr19_-_51472816
|
0.230
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr11_+_5968576
|
0.230
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chrX_-_109683457
|
0.229
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr20_+_51589752
|
0.228
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr13_+_50025687
|
0.228
|
NM_001160308
|
SETDB2
|
SET domain, bifurcated 2
|
|
chrX_-_76937157
|
0.227
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr3_-_114343791
|
0.226
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr4_+_88571453
|
0.224
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr11_-_121986800
|
0.223
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr12_-_111893857
|
0.222
|
|
|
|
|
chr7_-_140302311
|
0.220
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr2_-_158182154
|
0.220
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chrX_-_48327739
|
0.218
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr18_-_3219986
|
0.218
|
NM_003803
NM_019856
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr12_+_75874512
|
0.218
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chrX_+_100805415
|
0.217
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr22_+_20861964
|
0.217
|
|
MED15
|
mediator complex subunit 15
|
|
chr19_-_56249739
|
0.217
|
NM_176820
|
NLRP9
|
NLR family, pyrin domain containing 9
|
|
chr1_+_59775749
|
0.215
|
NM_001244714
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr9_-_34729463
|
0.212
|
NM_001141917
|
FAM205A
|
family with sequence similarity 205, member A
|
|
chr19_+_17326196
|
0.212
|
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr9_+_116207010
|
0.212
|
NM_144488
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr11_-_112131582
|
0.212
|
NM_001145024
|
C11orf34
|
chromosome 11 open reading frame 34
|
|
chr12_-_57504088
|
0.207
|
NM_001178079
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr8_+_92261515
|
0.206
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr16_-_1821503
|
0.205
|
|
NME3
|
non-metastatic cells 3, protein expressed in
|
|
chr6_-_79675672
|
0.205
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr11_-_84028379
|
0.203
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr12_-_10542616
|
0.203
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr11_-_31832758
|
0.203
|
|
PAX6
|
paired box 6
|