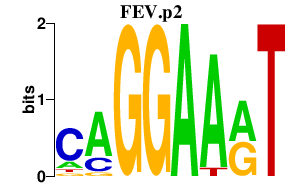
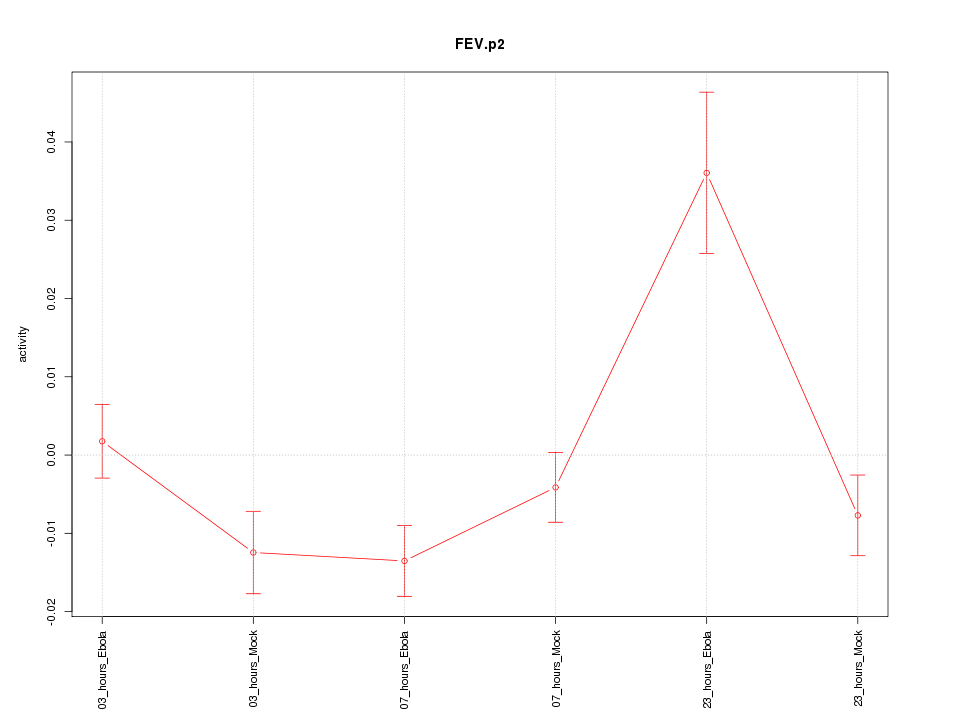
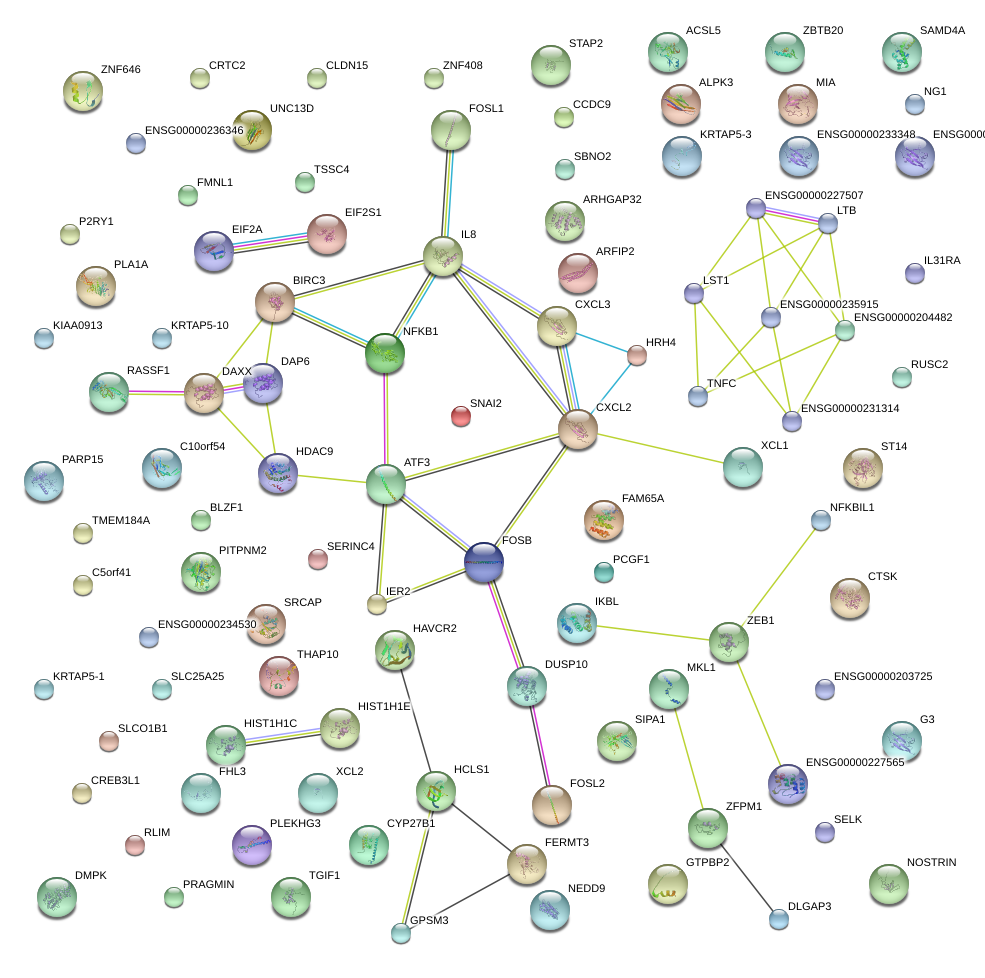
|
chr7_+_18535351
|
3.251
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_18548899
|
3.124
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_18548963
|
2.569
|
|
HDAC9
|
histone deacetylase 9
|
|
chr6_-_11382501
|
2.507
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_+_212738675
|
2.413
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr6_+_31515352
|
2.257
|
NM_001144961
NM_005007
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr2_+_169659099
|
2.207
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr3_-_114343052
|
2.185
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_-_8175860
|
2.043
|
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr17_+_43299277
|
1.941
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr6_-_32160289
|
1.863
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr7_+_18535817
|
1.790
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr11_+_46722564
|
1.719
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr7_-_100882100
|
1.687
|
NM_001185080
|
CLDN15
|
claudin 15
|
|
chr1_-_150780705
|
1.659
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr11_-_1606480
|
1.646
|
NM_001005922
|
KRTAP5-1
|
keratin associated protein 5-1
|
|
chr1_-_221910801
|
1.577
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr14_+_55034633
|
1.564
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr3_+_119316694
|
1.560
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr5_+_55149149
|
1.559
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr6_+_31515391
|
1.538
|
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr11_-_65667860
|
1.525
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_+_13261272
|
1.521
|
NM_004907
|
IER2
|
immediate early response 2
|
|
chr15_-_71184611
|
1.476
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr11_+_46722316
|
1.463
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr19_+_47759802
|
1.461
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr10_+_114154522
|
1.458
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr19_+_41281281
|
1.442
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr11_+_2421983
|
1.438
|
|
TSSC4
|
tumor suppressing subtransferable candidate 4
|
|
chr14_+_65171149
|
1.430
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr2_+_28616360
|
1.419
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr4_-_74964836
|
1.414
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr19_+_47759730
|
1.407
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr12_-_58159386
|
1.390
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr9_+_130854127
|
1.387
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr8_-_49833823
|
1.381
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr14_+_55034329
|
1.373
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr16_+_31085722
|
1.368
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr6_-_31620142
|
1.356
|
NM_001098534
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr6_+_26156558
|
1.337
|
NM_005321
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr4_-_74904329
|
1.320
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr6_-_11232900
|
1.314
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_31620468
|
1.312
|
NM_001199697
NM_001199698
NM_004639
NM_080702
NM_080703
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr19_+_45971249
|
1.298
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr14_+_65171243
|
1.297
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr16_+_31085751
|
1.290
|
|
ZNF646
|
zinc finger protein 646
|
|
chr18_+_22040592
|
1.290
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr11_+_63974190
|
1.287
|
|
FERMT3
|
fermitin family member 3
|
|
chr22_-_40859423
|
1.285
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr16_+_30710461
|
1.268
|
NM_006662
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chr10_-_73533196
|
1.254
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr19_-_1174201
|
1.244
|
|
|
|
|
chr2_+_28616346
|
1.229
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr4_+_103422553
|
1.225
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr5_-_132272803
|
1.223
|
|
|
|
|
chr1_-_153931096
|
1.214
|
NM_181715
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr15_+_85359910
|
1.213
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr1_+_168545710
|
1.210
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr1_-_153930911
|
1.197
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr6_-_31620426
|
1.195
|
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr6_-_31620384
|
1.192
|
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr6_+_31553955
|
1.191
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr2_-_74734498
|
1.187
|
|
PCGF1
|
polycomb group ring finger 1
|
|
chr3_+_150264514
|
1.174
|
NM_032025
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr6_-_43595046
|
1.161
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr11_-_6502563
|
1.155
|
NM_001242854
NM_001242855
NM_001242856
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr3_+_122334461
|
1.148
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr1_+_169337193
|
1.145
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr3_+_152552735
|
1.139
|
NM_002563
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1
|
|
chr10_+_75545363
|
1.138
|
NM_001242487
NM_001242488
NM_015037
|
KIAA0913
|
KIAA0913
|
|
chr11_+_102188180
|
1.133
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr15_-_44092134
|
1.122
|
NM_001033517
|
SERINC4
|
serine incorporator 4
|
|
chr19_-_1174257
|
1.120
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr19_-_46285752
|
1.118
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr16_+_67571980
|
1.118
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr3_-_50374749
|
1.117
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr1_-_153931077
|
1.113
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr9_+_35560424
|
1.113
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr1_+_156698270
|
1.112
|
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chr4_+_74606222
|
1.112
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr18_+_3453771
|
1.109
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr19_-_46285645
|
1.108
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr10_+_31610063
|
1.107
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr11_+_130029773
|
1.103
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr6_-_43595079
|
1.102
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr19_-_1174263
|
1.097
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr1_-_35370983
|
1.095
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr6_-_33290678
|
1.077
|
NM_001141969
NM_001141970
NM_001350
|
DAXX
|
death-domain associated protein
|
|
chr12_-_123590045
|
1.071
|
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr5_-_156536036
|
1.060
|
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chrX_-_73834460
|
1.058
|
NM_016120
NM_183353
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr11_+_46299585
|
1.041
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr5_+_172483285
|
1.041
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr11_-_128894008
|
1.041
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_+_169337432
|
1.033
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr1_+_156698250
|
1.033
|
NM_001142560
NM_015997
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chr7_-_1595746
|
1.032
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr3_-_121379701
|
1.022
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr1_-_38471170
|
1.013
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr6_-_31550065
|
1.012
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr19_-_4338750
|
1.012
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr11_+_65407591
|
1.010
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr3_-_53925871
|
0.999
|
NM_021237
|
SELK
|
selenoprotein K
|
|
chr14_+_77924415
|
0.998
|
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast)
|
|
chr5_-_43412417
|
0.998
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr19_-_55720812
|
0.989
|
NM_001161440
NM_002842
|
PTPRH
|
protein tyrosine phosphatase, receptor type, H
|
|
chr1_-_214724565
|
0.986
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr15_+_93443559
|
0.985
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr6_-_154677868
|
0.984
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr6_-_153323787
|
0.983
|
|
MTRF1L
|
mitochondrial translational release factor 1-like
|
|
chr4_+_88571453
|
0.983
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr7_-_99756275
|
0.982
|
NM_018275
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr2_+_28615695
|
0.979
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr15_+_93443418
|
0.974
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr4_+_186317837
|
0.971
|
NM_181726
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr4_+_103422822
|
0.970
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr17_-_62777621
|
0.968
|
|
LOC146880
|
Rho GTPase activating protein 27 pseudogene
|
|
chr1_+_155247186
|
0.963
|
NM_020897
|
HCN3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3
|
|
chr3_-_53381533
|
0.958
|
NM_018403
|
DCP1A
|
DCP1 decapping enzyme homolog A (S. cerevisiae)
|
|
chr9_-_130186491
|
0.954
|
|
|
|
|
chrX_-_48937560
|
0.954
|
NM_001029896
|
WDR45
|
WD repeat domain 45
|
|
chr7_+_76178604
|
0.952
|
|
LOC100133091
|
uncharacterized LOC100133091
|
|
chr4_-_74964996
|
0.949
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr9_-_14722669
|
0.949
|
NM_005454
|
CER1
|
cerberus 1, cysteine knot superfamily, homolog (Xenopus laevis)
|
|
chr2_+_37571752
|
0.945
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr4_-_184243578
|
0.942
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr19_-_56632600
|
0.942
|
|
ZNF787
|
zinc finger protein 787
|
|
chr6_+_26156552
|
0.930
|
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr7_+_157129659
|
0.926
|
NM_005494
NM_058246
|
DNAJB6
TMEM135
|
DnaJ (Hsp40) homolog, subfamily B, member 6
transmembrane protein 135
|
|
chr15_+_44092618
|
0.925
|
NM_001199885
NM_016400
|
C15orf63
|
chromosome 15 open reading frame 63
|
|
chr8_-_124665165
|
0.920
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr2_+_28615771
|
0.917
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_29038862
|
0.917
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr14_+_77924367
|
0.913
|
NM_012111
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast)
|
|
chr14_+_75745480
|
0.909
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr2_-_74734785
|
0.909
|
NM_032673
|
PCGF1
|
polycomb group ring finger 1
|
|
chr6_-_143266283
|
0.907
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr10_-_99160987
|
0.907
|
|
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae)
|
|
chr11_+_46299554
|
0.904
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr16_-_30366681
|
0.904
|
NM_006110
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2
|
|
chr19_+_15218141
|
0.903
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr17_-_72968750
|
0.897
|
|
|
|
|
chr17_-_72968790
|
0.894
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr2_+_28615668
|
0.890
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr4_-_54930706
|
0.889
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr5_-_176037091
|
0.887
|
NM_052899
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr15_+_41136622
|
0.884
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr17_-_38256548
|
0.879
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr14_-_21567069
|
0.876
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chr19_-_11616395
|
0.873
|
|
ZNF653
|
zinc finger protein 653
|
|
chr10_-_99161013
|
0.872
|
NM_001145114
NM_015179
|
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae)
|
|
chr2_-_69870976
|
0.869
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr20_-_44516237
|
0.867
|
NM_080608
|
SPATA25
|
spermatogenesis associated 25
|
|
chr4_+_103422730
|
0.860
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr17_-_29641094
|
0.860
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr19_+_16296211
|
0.858
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr22_-_37640287
|
0.857
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr4_+_103422479
|
0.857
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr14_+_102027687
|
0.857
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr19_+_48248798
|
0.854
|
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr9_-_4152182
|
0.852
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr19_-_46285746
|
0.849
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr17_+_62502860
|
0.849
|
|
CEP95
|
centrosomal protein 95kDa
|
|
chr1_+_68297985
|
0.846
|
|
LOC100289178
|
uncharacterized LOC100289178
|
|
chr19_-_59070324
|
0.846
|
NM_003969
|
UBE2M
|
ubiquitin-conjugating enzyme E2M
|
|
chr4_+_72897520
|
0.844
|
NM_004885
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chrX_-_48937457
|
0.844
|
|
WDR45
|
WD repeat domain 45
|
|
chr6_-_153323871
|
0.844
|
NM_001114184
NM_019041
|
MTRF1L
|
mitochondrial translational release factor 1-like
|
|
chr19_+_48248792
|
0.839
|
NM_015710
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr11_-_72433379
|
0.836
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr17_-_38256905
|
0.835
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr19_-_6279797
|
0.835
|
|
|
|
|
chr1_+_182569427
|
0.834
|
|
|
|
|
chr7_-_99756329
|
0.834
|
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr8_+_145582208
|
0.829
|
NM_001253816
NM_001253815
|
GPR172A
|
G protein-coupled receptor 172A
|
|
chr12_-_57914287
|
0.827
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr1_+_201979689
|
0.826
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr15_-_80263460
|
0.826
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr10_+_75545296
|
0.822
|
|
KIAA0913
|
KIAA0913
|
|
chr14_+_23938883
|
0.818
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr3_+_150264576
|
0.818
|
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr6_-_108582307
|
0.817
|
NM_003795
NM_152827
|
SNX3
|
sorting nexin 3
|
|
chr4_-_54930466
|
0.816
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr11_+_47290893
|
0.815
|
NM_130470
|
MADD
|
MAP-kinase activating death domain
|
|
chr12_+_7053180
|
0.815
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr7_+_92076762
|
0.814
|
NM_021167
|
GATAD1
|
GATA zinc finger domain containing 1
|
|
chr1_-_154946830
|
0.813
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr7_-_16505292
|
0.811
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr11_+_61129831
|
0.808
|
|
TMEM138
|
transmembrane protein 138
|
|
chr4_+_110736911
|
0.808
|
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr14_+_67999915
|
0.806
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr16_+_30773619
|
0.806
|
|
RNF40
|
ring finger protein 40
|
|
chr17_+_16318848
|
0.806
|
NM_016113
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr9_+_139971938
|
0.803
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr6_-_31648140
|
0.803
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr8_-_79717540
|
0.803
|
NM_000880
NM_001199886
NM_001199887
NM_001199888
|
IL7
|
interleukin 7
|
|
chr21_+_30517897
|
0.801
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr2_+_148778573
|
0.800
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr8_-_42234562
|
0.798
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr1_-_156542320
|
0.796
|
NM_178229
|
IQGAP3
|
IQ motif containing GTPase activating protein 3
|