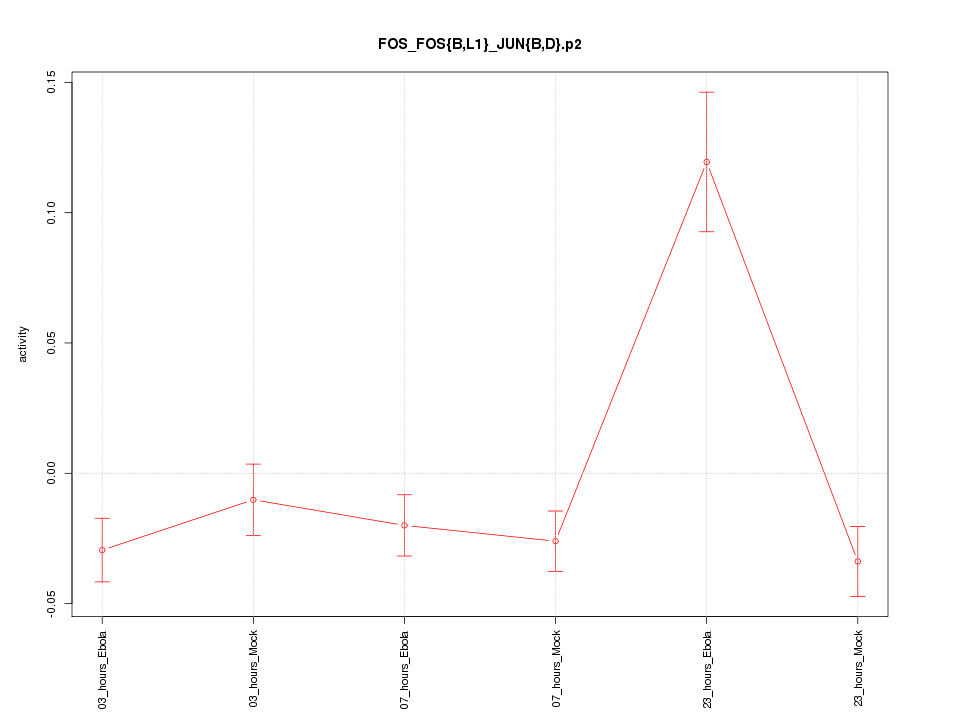
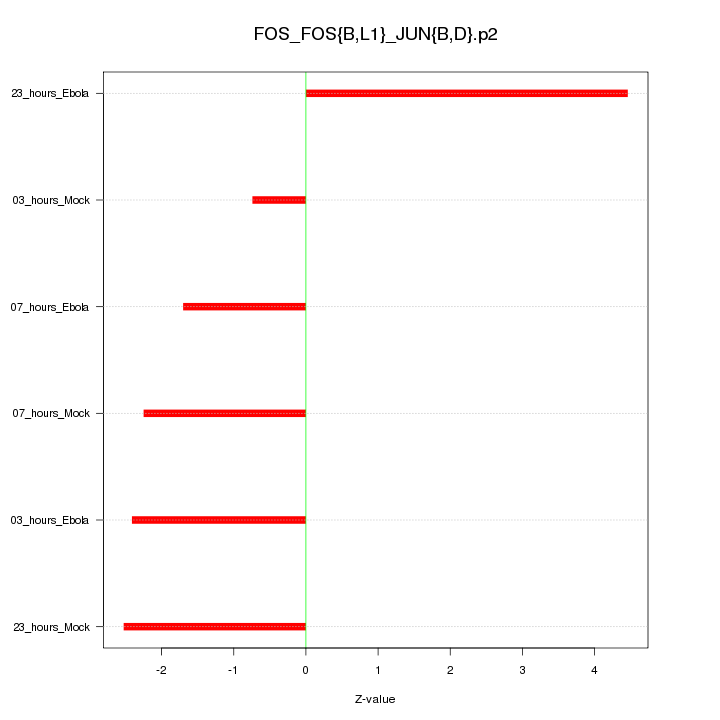
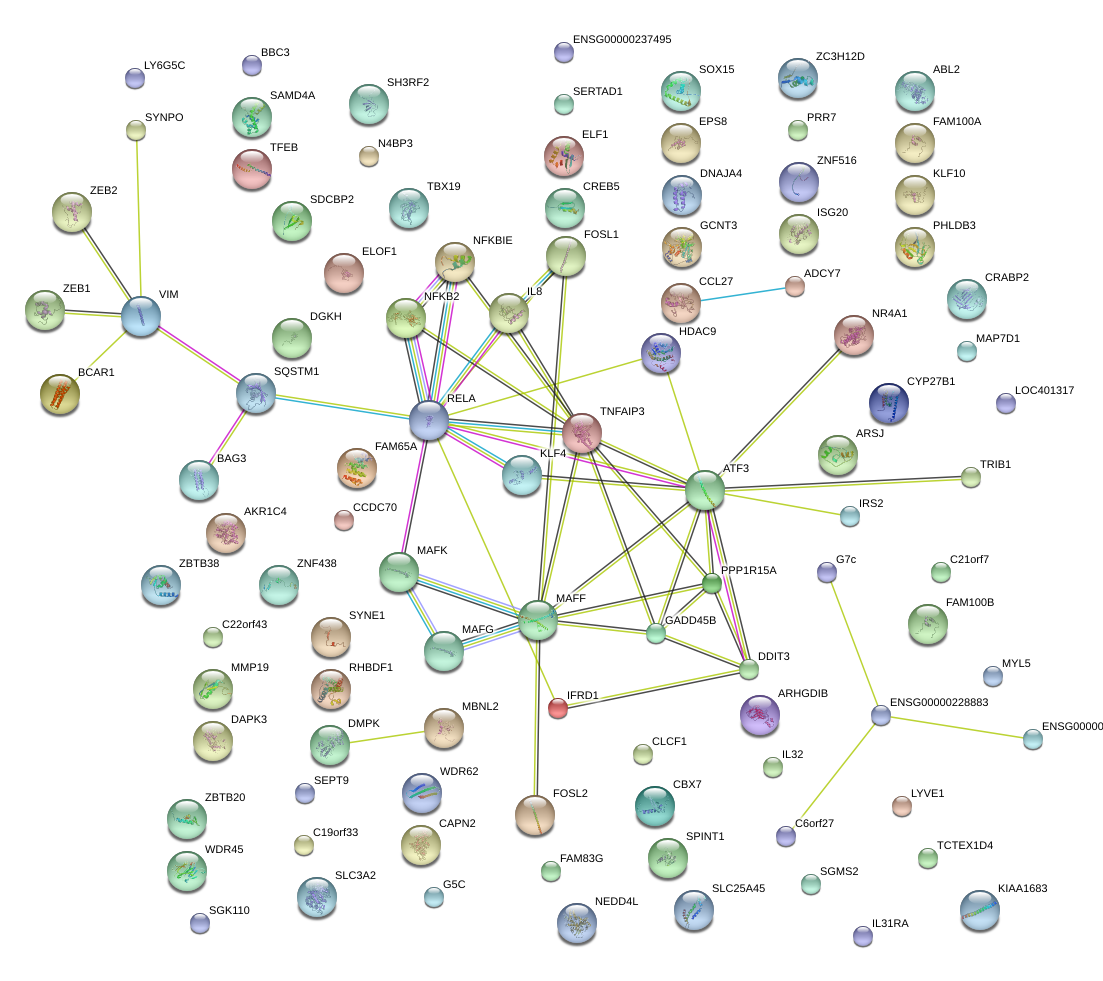
|
chr12_-_58159386
|
9.565
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr4_+_74606222
|
8.086
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr19_-_56056908
|
7.271
|
NM_001199824
|
SGK110
|
putative uncharacterized serine/threonine-protein kinase SgK110-like
|
|
chr19_-_47735858
|
7.210
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr5_+_176875325
|
7.067
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr1_+_223889253
|
6.992
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr19_-_46285645
|
6.697
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr1_-_45272956
|
5.869
|
NM_001013632
|
TCTEX1D4
|
Tctex1 domain containing 4
|
|
chr19_+_49376488
|
5.847
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr13_+_97794050
|
5.596
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_-_65667809
|
5.194
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr7_+_1577118
|
5.055
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr13_-_41593405
|
5.010
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr9_-_34662629
|
4.948
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr7_+_28725597
|
4.900
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_150020201
|
4.805
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr2_+_28615771
|
4.537
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr17_-_18907988
|
4.479
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr13_+_52436116
|
4.448
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr19_-_40931880
|
4.401
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr6_-_152702450
|
4.303
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr2_+_87754991
|
4.232
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr7_+_28448970
|
4.095
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr16_-_75301950
|
4.070
|
NM_001170715
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr19_-_36019194
|
3.986
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr13_-_110438896
|
3.985
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr1_+_212738675
|
3.937
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr1_-_179112178
|
3.781
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr21_+_30517897
|
3.725
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr18_+_55888750
|
3.723
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55888838
|
3.683
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_+_87755059
|
3.607
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr11_-_65667860
|
3.582
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr11_-_65150107
|
3.492
|
NM_001077241
NM_182556
|
SLC25A45
|
solute carrier family 25, member 45
|
|
chr2_+_28615695
|
3.486
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr13_-_41593491
|
3.469
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr6_-_44233499
|
3.465
|
NM_004556
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr6_-_31648140
|
3.458
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr19_-_46285752
|
3.447
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr18_+_55888799
|
3.331
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_-_44233244
|
3.302
|
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr7_+_18535351
|
3.213
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr4_-_114900802
|
3.135
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr19_-_3969734
|
3.133
|
NM_001348
|
DAPK3
|
death-associated protein kinase 3
|
|
chr12_-_57914287
|
3.111
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr18_-_74207145
|
3.075
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr2_-_145274916
|
3.056
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr6_-_41703938
|
3.019
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr16_-_122573
|
2.998
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr11_-_67141647
|
2.994
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr4_+_108745718
|
2.989
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr1_-_156675237
|
2.958
|
NM_001878
NM_001199723
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr17_-_7493389
|
2.919
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr10_+_5238756
|
2.894
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr8_-_103666018
|
2.868
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr7_+_112063131
|
2.853
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr12_-_56236614
|
2.845
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr3_-_114343791
|
2.795
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr19_-_44008984
|
2.787
|
NM_198850
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3
|
|
chr13_+_97928414
|
2.781
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr22_+_38597938
|
2.766
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr19_-_18385166
|
2.760
|
NM_001145304
NM_001145305
NM_025249
|
KIAA1683
|
KIAA1683
|
|
chr15_+_89182038
|
2.757
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr22_+_38598065
|
2.756
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr14_+_55168831
|
2.678
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr12_-_15114495
|
2.657
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr19_+_38794800
|
2.645
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr5_+_55149149
|
2.642
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr11_+_62649160
|
2.635
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr16_-_4665002
|
2.604
|
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr11_-_65430427
|
2.599
|
NM_001145138
NM_001243984
NM_001243985
NM_021975
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr19_-_46285746
|
2.573
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr15_+_78558522
|
2.548
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr16_+_67571342
|
2.521
|
NM_001193524
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr1_+_223900328
|
2.513
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_+_223900226
|
2.509
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_+_223900212
|
2.508
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr10_+_17270469
|
2.504
|
|
VIM
|
vimentin
|
|
chr17_+_74261280
|
2.504
|
NM_182565
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr17_+_74261431
|
2.429
|
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr14_+_55221505
|
2.428
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr9_-_110252045
|
2.406
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr11_-_65430214
|
2.345
|
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr6_+_138188352
|
2.337
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr4_+_671710
|
2.336
|
NM_002477
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr3_+_141087298
|
2.333
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr5_+_177540465
|
2.320
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr5_+_179247841
|
2.312
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr5_+_179247903
|
2.305
|
|
SQSTM1
|
sequestosome 1
|
|
chr2_+_28615668
|
2.298
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr13_+_97928196
|
2.291
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr22_-_39529021
|
2.274
|
|
CBX7
|
chromobox homolog 7
|
|
chr17_-_79881304
|
2.264
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr8_+_126442557
|
2.247
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr5_+_179247911
|
2.230
|
|
SQSTM1
|
sequestosome 1
|
|
chr10_+_104155431
|
2.179
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr12_+_52445185
|
2.154
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr10_-_31288420
|
2.151
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr11_-_10590078
|
2.138
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr13_+_42712177
|
2.110
|
NM_001204505
NM_001204506
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr15_+_59903980
|
2.108
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr16_-_4664780
|
2.107
|
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chrX_-_48937560
|
2.102
|
NM_001029896
|
WDR45
|
WD repeat domain 45
|
|
chr22_-_23974486
|
2.098
|
NM_016449
|
C22orf43
|
chromosome 22 open reading frame 43
|
|
chr5_+_145317297
|
2.088
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr15_+_59904090
|
2.084
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr21_-_37528578
|
2.060
|
|
LOC100506428
|
uncharacterized LOC100506428
|
|
chr16_+_50300461
|
2.050
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr20_-_1309782
|
2.032
|
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr1_+_168250277
|
2.031
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr19_+_2476122
|
2.008
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr16_+_3115639
|
1.995
|
|
IL32
|
interleukin 32
|
|
chr12_-_15815073
|
1.993
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr1_+_36621751
|
1.988
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr6_-_149806116
|
1.945
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr15_+_41136178
|
1.936
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr13_+_51915167
|
1.929
|
NM_001101320
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3
|
|
chr10_+_121410751
|
1.912
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr6_-_31745048
|
1.903
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr16_-_75272524
|
1.898
|
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr1_-_244006471
|
1.891
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chrX_-_48937457
|
1.872
|
|
WDR45
|
WD repeat domain 45
|
|
chr2_-_73316782
|
1.865
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr11_-_65667858
|
1.859
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr12_+_121416294
|
1.853
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr15_+_41136622
|
1.850
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr7_-_100881030
|
1.850
|
NM_014343
|
CLDN15
|
claudin 15
|
|
chr15_+_86220267
|
1.844
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chrX_+_153770458
|
1.833
|
NM_001099856
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr3_+_160394946
|
1.830
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr11_-_128894008
|
1.818
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr17_+_79651072
|
1.802
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr10_+_121410936
|
1.787
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr6_-_41673677
|
1.787
|
|
TFEB
|
transcription factor EB
|
|
chr10_+_80828776
|
1.780
|
NM_020338
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr10_+_17270257
|
1.776
|
NM_003380
|
VIM
|
vimentin
|
|
chr17_-_17399494
|
1.768
|
NM_001199989
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr6_-_152702329
|
1.765
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_+_103191795
|
1.761
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr16_+_53133063
|
1.752
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr10_+_121410891
|
1.740
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr1_+_168545710
|
1.711
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr16_+_67563539
|
1.709
|
NM_001193523
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr13_-_101185944
|
1.702
|
NM_033110
|
A2LD1
|
AIG2-like domain 1
|
|
chr17_-_33390676
|
1.691
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr9_-_139890984
|
1.687
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr12_-_39299335
|
1.677
|
NM_153634
|
CPNE8
|
copine VIII
|
|
chr3_-_187454247
|
1.665
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr17_+_79651035
|
1.643
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr13_-_30423599
|
1.632
|
|
UBL3
|
ubiquitin-like 3
|
|
chr16_-_4664894
|
1.627
|
NM_145253
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr14_-_23445885
|
1.607
|
|
AJUBA
|
ajuba LIM protein
|
|
chr6_-_46293215
|
1.580
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_+_156084458
|
1.574
|
NM_005572
NM_170707
NM_170708
|
LMNA
|
lamin A/C
|
|
chr2_-_85641128
|
1.569
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr2_-_202563353
|
1.569
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr3_+_49027281
|
1.566
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr17_-_8022134
|
1.565
|
NM_001165960
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr7_+_48128225
|
1.564
|
NM_181597
|
UPP1
|
uridine phosphorylase 1
|
|
chr6_+_12290528
|
1.557
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr17_+_27573874
|
1.552
|
NM_005208
|
CRYBA1
|
crystallin, beta A1
|
|
chr19_-_1132109
|
1.542
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr6_+_33387846
|
1.533
|
NM_006772
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr21_-_43735462
|
1.517
|
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr1_+_36621457
|
1.510
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr2_-_145275086
|
1.485
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_-_27816542
|
1.482
|
|
WASF2
|
WAS protein family, member 2
|
|
chr11_+_63655986
|
1.468
|
NM_017490
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr9_+_74920329
|
1.465
|
|
|
|
|
chr15_-_56209305
|
1.465
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr4_+_108814419
|
1.459
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr17_+_79650961
|
1.451
|
NM_004712
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr11_+_62475113
|
1.439
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr17_+_79651042
|
1.437
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr6_-_88854989
|
1.431
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr19_+_7953380
|
1.417
|
NM_025061
|
LRRC8E
|
leucine rich repeat containing 8 family, member E
|
|
chr10_+_121411086
|
1.404
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr3_-_99833348
|
1.393
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr14_+_68168602
|
1.384
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr3_+_141043054
|
1.371
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr8_+_22210311
|
1.364
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr13_-_60325073
|
1.364
|
|
|
|
|
chr7_+_127233688
|
1.363
|
NM_020369
|
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus)
|
|
chr7_+_39017608
|
1.361
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr9_-_35111534
|
1.355
|
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr1_-_27816667
|
1.354
|
|
WASF2
|
WAS protein family, member 2
|
|
chr1_-_27816676
|
1.351
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr2_+_219125737
|
1.345
|
NM_001077191
NM_001077194
NM_170699
|
GPBAR1
|
G protein-coupled bile acid receptor 1
|
|
chr2_-_208994408
|
1.326
|
NM_020989
|
CRYGC
|
crystallin, gamma C
|
|
chr10_+_104154228
|
1.321
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr2_+_30369749
|
1.317
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chrX_-_48900752
|
1.313
|
|
TFE3
|
transcription factor binding to IGHM enhancer 3
|
|
chr22_+_20861875
|
1.308
|
|
MED15
|
mediator complex subunit 15
|
|
chr3_-_87325611
|
1.300
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr2_+_33359607
|
1.298
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr1_+_178063631
|
1.293
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr13_+_97874540
|
1.292
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_+_145316120
|
1.290
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr10_-_76868853
|
1.287
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr16_+_30419363
|
1.282
|
|
ZNF771
|
zinc finger protein 771
|