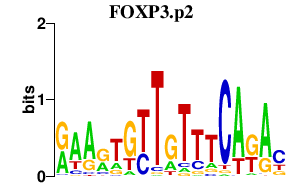
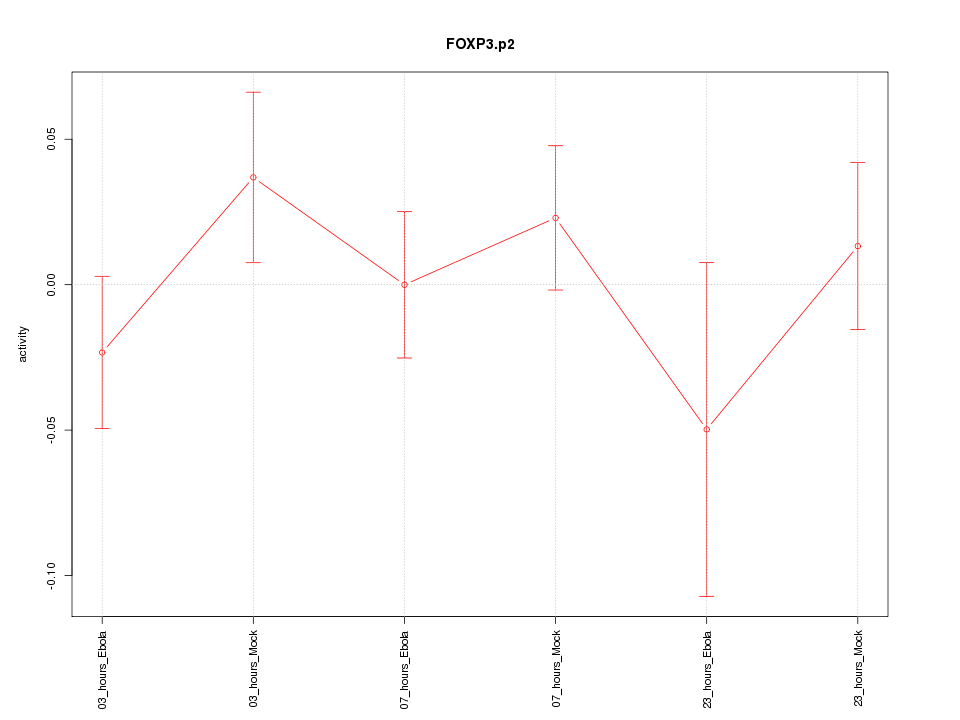
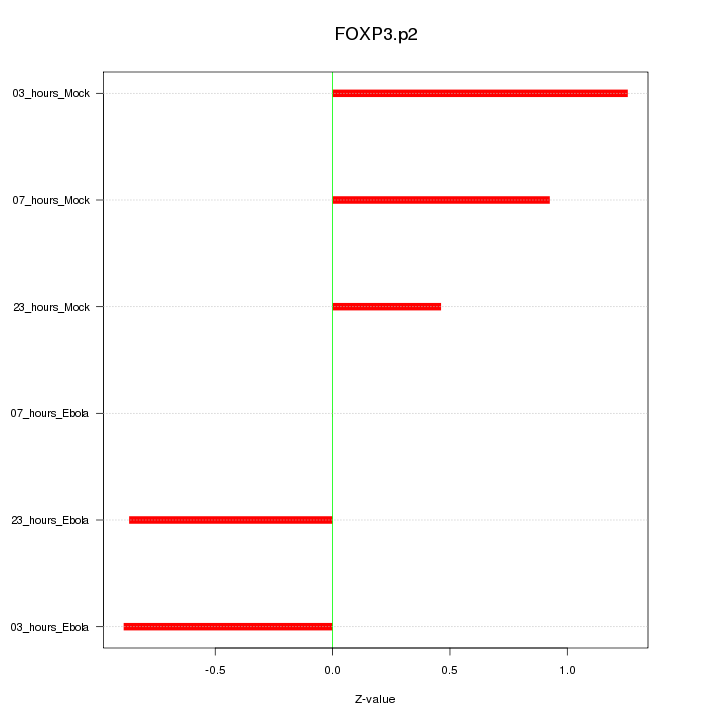
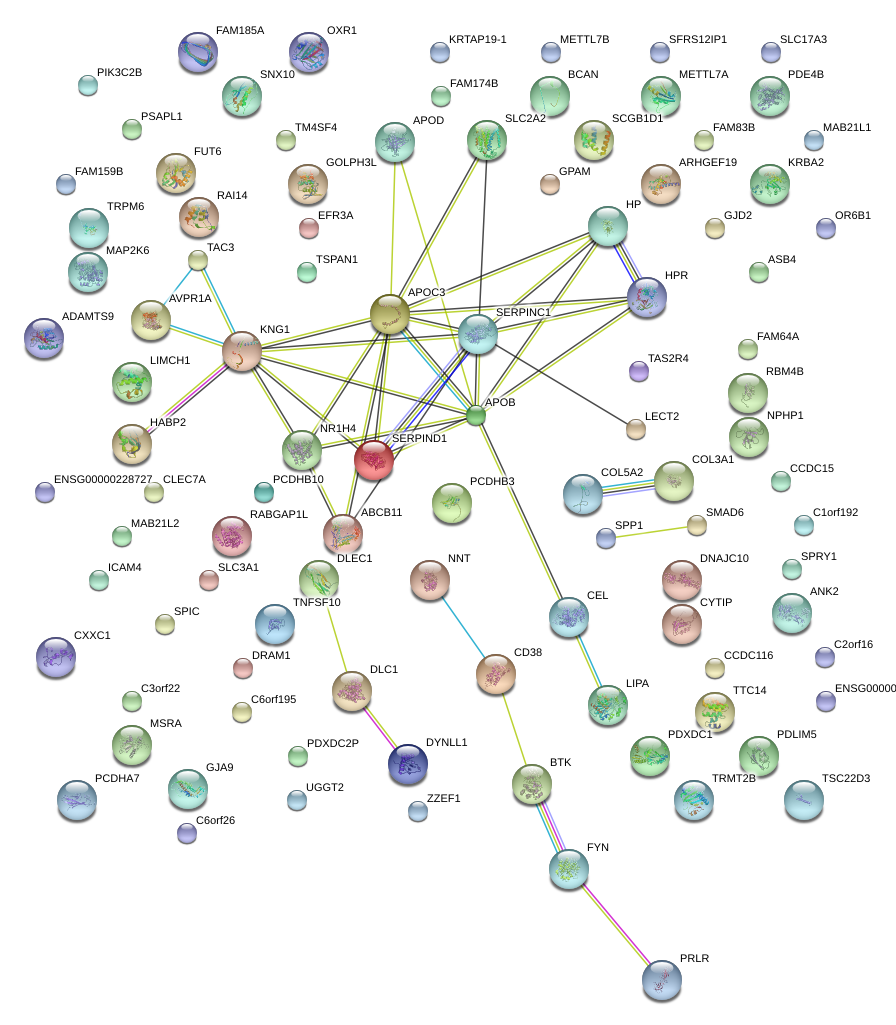
|
chr2_+_65663812
|
1.482
|
|
FLJ16124
|
FLJ16124 protein
|
|
chr16_+_72088507
|
1.349
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr11_+_124824076
|
0.887
|
|
CCDC15
|
coiled-coil domain containing 15
|
|
chr6_+_31730772
|
0.850
|
NM_001039651
|
C6orf26
|
chromosome 6 open reading frame 26
|
|
chr1_-_16539103
|
0.818
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr16_+_72097124
|
0.791
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr22_+_21128378
|
0.766
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr11_+_124824012
|
0.762
|
NM_025004
|
CCDC15
|
coiled-coil domain containing 15
|
|
chr5_+_140213968
|
0.727
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr2_-_21228189
|
0.701
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr7_+_95115212
|
0.701
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr5_+_140782367
|
0.689
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr17_-_8274852
|
0.560
|
NM_213597
|
KRBA2
|
KRAB-A domain containing 2
|
|
chr4_+_15779887
|
0.548
|
NM_001775
|
CD38
|
CD38 molecule
|
|
chr17_+_67410825
|
0.528
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr4_+_114214102
|
0.480
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr10_-_91102430
|
0.476
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr4_+_88896801
|
0.464
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr6_-_25874439
|
0.459
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr11_+_116700620
|
0.459
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr2_-_158300517
|
0.443
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr12_-_10282680
|
0.395
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chrX_-_106960268
|
0.387
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_15990300
|
0.369
|
|
|
|
|
chr5_+_140743755
|
0.369
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr16_+_15068921
|
0.356
|
|
PDXDC1
|
pyridoxal-dependent decarboxylase domain containing 1
|
|
chr5_-_135290521
|
0.344
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr1_-_150669499
|
0.337
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr5_+_63986134
|
0.328
|
NM_001164442
|
FAM159B
|
family with sequence similarity 159, member B
|
|
chr7_+_143701021
|
0.320
|
NM_001005281
|
OR6B1
|
olfactory receptor, family 6, subfamily B, member 1
|
|
chr8_+_32785485
|
0.320
|
|
|
|
|
chr3_-_172241060
|
0.318
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chr1_+_174844655
|
0.311
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr21_-_31852631
|
0.310
|
NM_181607
|
KRTAP19-1
|
keratin associated protein 19-1
|
|
chr4_+_88896838
|
0.306
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr10_-_113933517
|
0.304
|
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr12_+_100867739
|
0.302
|
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr12_+_100897137
|
0.291
|
NM_001206992
NM_001206993
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr2_+_44502596
|
0.286
|
NM_000341
|
SLC3A1
|
solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1
|
|
chr3_-_170744458
|
0.278
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr4_+_88896899
|
0.278
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr8_+_9953061
|
0.276
|
NM_001135671
NM_001199729
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr2_-_21225640
|
0.269
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr8_+_107738239
|
0.265
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr12_+_51318515
|
0.265
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr2_-_169887832
|
0.262
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr5_-_35118223
|
0.254
|
NM_001204314
NM_001204315
NM_001204316
NM_001204317
NM_001204318
|
PRLR
|
prolactin receptor
|
|
chr1_+_244217283
|
0.244
|
|
|
|
|
chr2_+_189839132
|
0.240
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr3_-_64672285
|
0.239
|
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr2_-_190044330
|
0.233
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr1_-_39347258
|
0.229
|
NM_030772
|
GJA9-MYCBP
GJA9
|
GJA9-MYCBP readthrough
gap junction protein, alpha 9, 59kDa
|
|
chr15_+_67000929
|
0.227
|
NM_001142861
|
SMAD6
|
SMAD family member 6
|
|
chr2_-_110962539
|
0.221
|
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr17_+_6347733
|
0.221
|
NM_001195228
NM_019013
|
FAM64A
|
family with sequence similarity 64, member A
|
|
chr22_+_21986968
|
0.216
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr5_+_140566655
|
0.216
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr15_-_93277303
|
0.215
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chrX_-_100307042
|
0.215
|
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr4_+_151503076
|
0.214
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr4_+_41540187
|
0.210
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr12_-_57410275
|
0.209
|
NM_001178054
NM_013251
|
TAC3
|
tachykinin 3
|
|
chr5_+_140571903
|
0.208
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr7_+_26400503
|
0.208
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr16_+_15068832
|
0.204
|
NM_015027
|
PDXDC1
|
pyridoxal-dependent decarboxylase domain containing 1
|
|
chr5_+_140480056
|
0.204
|
NM_018937
|
PCDHB3
|
protocadherin beta 3
|
|
chr10_+_115310589
|
0.192
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr12_+_56075418
|
0.190
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr8_+_132991171
|
0.188
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr12_+_100867550
|
0.185
|
NM_001206977
NM_001206978
NM_001206979
NM_005123
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr7_+_102389325
|
0.185
|
NM_001145268
NM_001145269
|
FAM185A
|
family with sequence similarity 185, member A
|
|
chr6_+_132859426
|
0.169
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr3_+_149192479
|
0.157
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr9_-_77503009
|
0.153
|
NM_017662
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr3_-_126277757
|
0.149
|
NM_152533
|
C3orf22
|
chromosome 3 open reading frame 22
|
|
chr7_+_141478241
|
0.147
|
NM_016944
|
TAS2R4
|
taste receptor, type 2, member 4
|
|
chrX_-_100307091
|
0.147
|
NM_001167970
NM_001167971
NM_001167972
NM_024917
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr3_+_186435097
|
0.147
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr4_+_95376396
|
0.145
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr4_+_124317884
|
0.145
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_+_46646199
|
0.144
|
|
TSPAN1
|
tetraspanin 1
|
|
chr5_+_43602738
|
0.143
|
NM_012343
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr3_+_180320955
|
0.141
|
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr12_-_63546589
|
0.140
|
NM_000706
|
AVPR1A
|
arginine vasopressin receptor 1A
|
|
chr1_-_204459445
|
0.139
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr4_-_7436614
|
0.139
|
NM_001085382
|
PSAPL1
|
prosaposin-like 1 (gene/pseudogene)
|
|
chr9_+_686640
|
0.135
|
|
|
|
|
chr1_-_198906536
|
0.134
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr6_-_2635297
|
0.133
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr18_+_3594056
|
0.133
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr3_-_195310770
|
0.131
|
|
APOD
|
apolipoprotein D
|
|
chr5_+_34687663
|
0.131
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr6_-_112041253
|
0.131
|
NM_153047
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr1_-_173886464
|
0.131
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr1_+_66798095
|
0.131
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr19_-_5839702
|
0.131
|
NM_000150
NM_001040701
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr2_-_110962577
|
0.130
|
NM_000272
NM_001128178
NM_001128179
NM_207181
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr13_-_96705565
|
0.129
|
NM_020121
|
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2
|
|
chr2_+_183608329
|
0.129
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr6_+_54711568
|
0.128
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr19_+_10397642
|
0.128
|
NM_001039132
NM_001544
NM_022377
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|
|
chr2_+_27799388
|
0.128
|
NM_032266
|
C2orf16
|
chromosome 2 open reading frame 16
|
|
chrX_-_100641143
|
0.126
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr11_-_66445218
|
0.125
|
NM_031492
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr12_+_102271349
|
0.123
|
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr1_-_161337663
|
0.121
|
NM_001013625
|
C1orf192
|
chromosome 1 open reading frame 192
|
|
chr1_+_227751260
|
0.120
|
|
ZNF678
|
zinc finger protein 678
|
|
chr3_-_149086884
|
0.118
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr7_-_107880492
|
0.117
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr4_-_186732047
|
0.116
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_156030965
|
0.116
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr1_+_196912910
|
0.114
|
NM_005666
|
CFHR2
|
complement factor H-related 2
|
|
chr9_+_132962871
|
0.114
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr7_+_123485222
|
0.114
|
NM_012269
|
HYAL4
|
hyaluronoglucosaminidase 4
|
|
chr16_-_12897632
|
0.112
|
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr3_+_49547867
|
0.110
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr1_-_78444690
|
0.105
|
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr4_+_47033294
|
0.105
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr4_-_164534656
|
0.103
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr1_+_244216506
|
0.103
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr2_+_198570027
|
0.102
|
NM_138395
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial
|
|
chr13_+_49684388
|
0.101
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr2_-_75796847
|
0.101
|
NM_032181
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr1_+_164818083
|
0.099
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr7_-_95064213
|
0.099
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr2_+_189839078
|
0.097
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr5_+_43609261
|
0.096
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr19_-_45579687
|
0.096
|
NM_145288
|
ZNF296
|
zinc finger protein 296
|
|
chr8_+_92114846
|
0.095
|
NM_001129890
|
LRRC69
|
leucine rich repeat containing 69
|
|
chr22_-_38881529
|
0.092
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr6_-_38563763
|
0.092
|
NM_001172418
NM_152733
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr4_-_186697065
|
0.091
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_78444772
|
0.089
|
NM_003902
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr1_+_66797775
|
0.088
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr12_-_8219001
|
0.088
|
|
C3AR1
|
complement component 3a receptor 1
|
|
chr8_-_25282555
|
0.087
|
NM_000825
NM_001083111
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone)
|
|
chr11_-_66445318
|
0.087
|
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr3_-_71778268
|
0.086
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr20_-_270957
|
0.084
|
|
C20orf96
|
chromosome 20 open reading frame 96
|
|
chrX_-_117119282
|
0.084
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr20_+_54933982
|
0.084
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr3_+_124103551
|
0.082
|
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr5_+_140501399
|
0.082
|
|
PCDHB4
|
protocadherin beta 4
|
|
chrX_-_131353460
|
0.082
|
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr8_+_120079423
|
0.082
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr3_+_149192367
|
0.082
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr8_-_90996785
|
0.081
|
|
NBN
|
nibrin
|
|
chr10_+_95517565
|
0.080
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr1_+_145487301
|
0.080
|
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr3_-_51533996
|
0.079
|
NM_001171904
NM_014703
|
VPRBP
|
Vpr (HIV-1) binding protein
|
|
chr6_+_42952326
|
0.079
|
|
PPP2R5D
|
protein phosphatase 2, regulatory subunit B', delta
|
|
chr5_+_40909617
|
0.078
|
|
C7
|
complement component 7
|
|
chr11_-_6790187
|
0.078
|
NM_001004490
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2
|
|
chr10_+_74653338
|
0.078
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr10_-_106093662
|
0.078
|
NM_033397
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein
|
|
chr10_-_50732142
|
0.077
|
NM_170753
|
PGBD3
|
piggyBac transposable element derived 3
|
|
chr2_+_234627423
|
0.076
|
NM_007120
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4
|
|
chr10_-_118680978
|
0.073
|
|
KIAA1598
|
KIAA1598
|
|
chr1_-_173886401
|
0.073
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chrX_+_102965836
|
0.073
|
NM_182541
|
TMEM31
|
transmembrane protein 31
|
|
chrX_+_49832214
|
0.073
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr15_+_45021319
|
0.072
|
|
TRIM69
|
tripartite motif containing 69
|
|
chr5_-_110062365
|
0.072
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr5_+_40909558
|
0.072
|
NM_000587
|
C7
|
complement component 7
|
|
chr8_-_81787015
|
0.072
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr1_-_109935869
|
0.072
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr5_+_61874561
|
0.071
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr17_+_9729380
|
0.071
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr11_-_8892909
|
0.071
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr9_-_100700422
|
0.070
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr6_+_64356430
|
0.070
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr3_-_42917191
|
0.067
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr1_+_47881743
|
0.067
|
NM_012186
|
FOXE3
|
forkhead box E3
|
|
chr19_+_4278597
|
0.067
|
NM_020209
|
SHD
|
Src homology 2 domain containing transforming protein D
|
|
chr8_-_37727937
|
0.067
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr11_-_47545539
|
0.066
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr2_+_211458078
|
0.066
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr8_-_62602285
|
0.066
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr4_-_103245468
|
0.065
|
NM_001135148
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr21_-_33957746
|
0.064
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr4_+_86699858
|
0.064
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_+_92632608
|
0.064
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr19_-_36303766
|
0.064
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr22_-_36236421
|
0.064
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr12_-_3982394
|
0.062
|
NM_020367
|
PARP11
|
poly (ADP-ribose) polymerase family, member 11
|
|
chr12_-_91505541
|
0.062
|
NM_002345
|
LUM
|
lumican
|
|
chr10_+_91092238
|
0.062
|
NM_001031683
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr2_+_170366211
|
0.061
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr19_-_47291594
|
0.061
|
NM_005628
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr1_-_153283166
|
0.061
|
NM_052891
|
PGLYRP3
|
peptidoglycan recognition protein 3
|
|
chr11_-_74204708
|
0.061
|
NM_001144869
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative)
|
|
chr3_-_124732379
|
0.060
|
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr2_-_74382352
|
0.060
|
|
|
|
|
chr16_-_12897693
|
0.059
|
NM_001099455
NM_018340
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr19_-_23578226
|
0.059
|
NM_003430
|
ZNF91
|
zinc finger protein 91
|
|
chr7_-_64467006
|
0.059
|
NM_001007253
|
ERV3-1
|
endogenous retrovirus group 3, member 1
|
|
chr1_-_115269672
|
0.058
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr2_-_114036487
|
0.058
|
NM_003466
NM_013952
NM_013953
NM_013992
|
PAX8
|
paired box 8
|
|
chr4_+_187070326
|
0.058
|
NM_001006655
|
FAM149A
|
family with sequence similarity 149, member A
|
|
chr3_-_170744529
|
0.057
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|