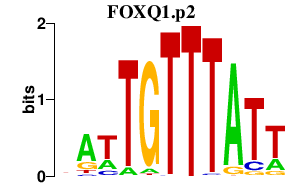
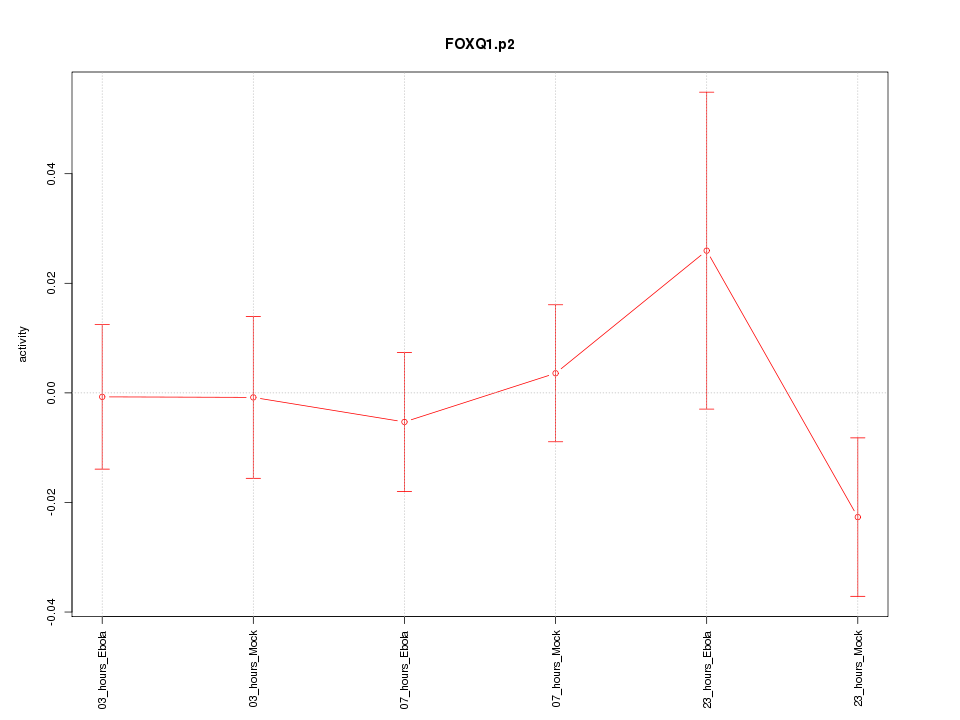
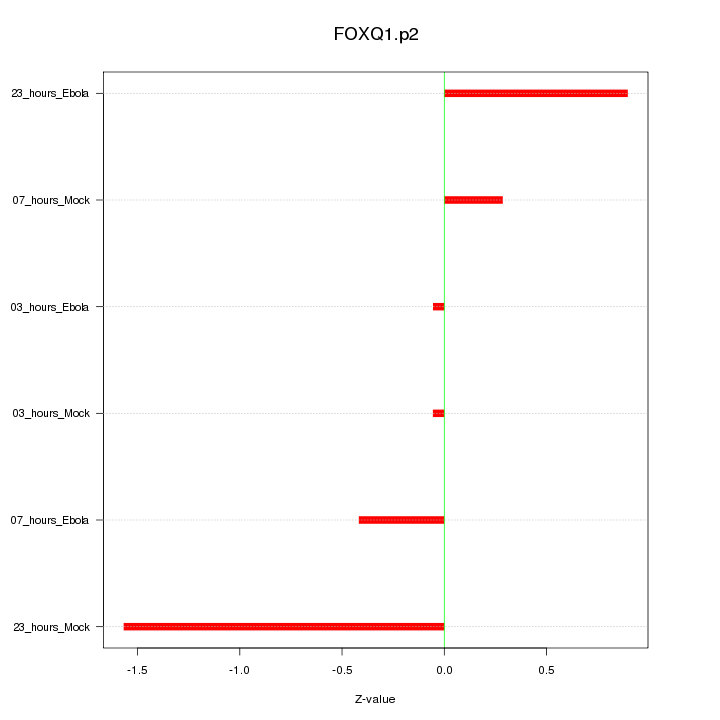
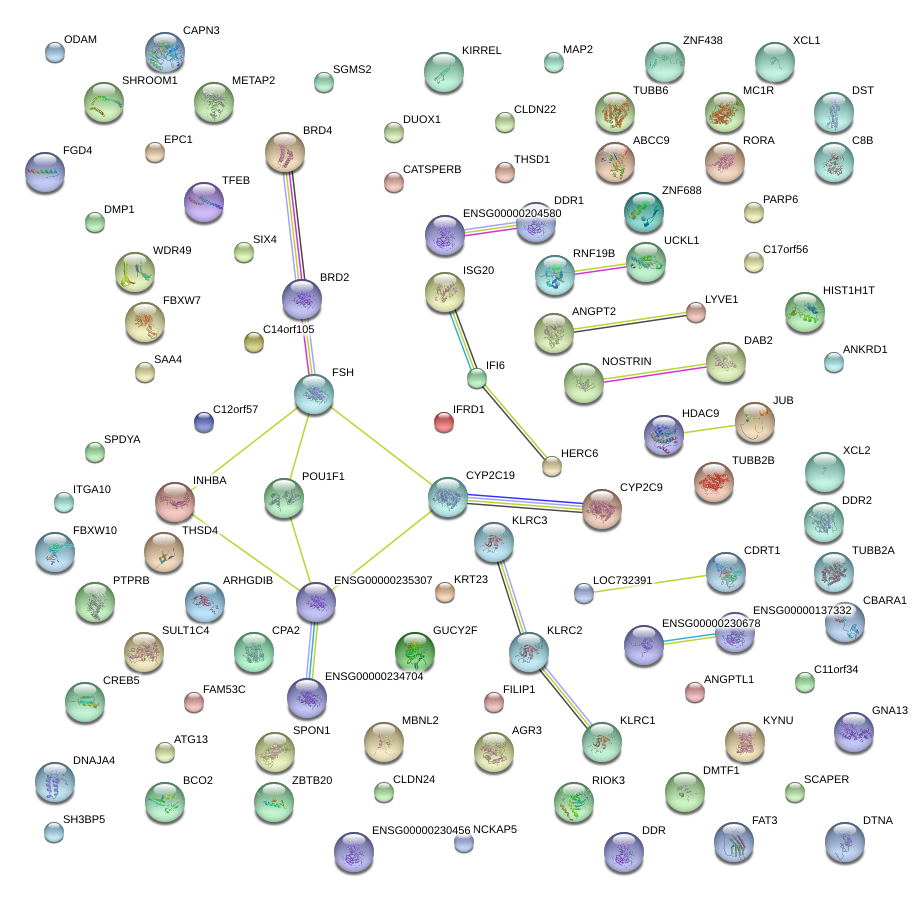
|
chrM_+_10123
|
1.475
|
|
|
|
|
chr7_+_28338939
|
1.437
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_28452143
|
1.436
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_15114495
|
0.952
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr12_-_22089627
|
0.850
|
NM_005691
NM_020297
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr15_+_71839540
|
0.817
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr3_-_114343052
|
0.808
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_18535817
|
0.786
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr7_-_41742666
|
0.780
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr17_-_63009302
|
0.726
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr18_+_32335939
|
0.721
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr2_+_29038862
|
0.673
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr6_-_26108320
|
0.668
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr4_-_184241926
|
0.593
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr10_-_92681025
|
0.585
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr11_-_112131582
|
0.581
|
NM_001145024
|
C11orf34
|
chromosome 11 open reading frame 34
|
|
chr1_+_168545710
|
0.575
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr3_-_114478117
|
0.557
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_18330044
|
0.547
|
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_28725597
|
0.514
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_-_41703938
|
0.513
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr2_+_108994366
|
0.513
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr4_+_71062241
|
0.496
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr18_+_21032786
|
0.488
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr3_-_15382874
|
0.483
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr5_-_39424930
|
0.476
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_-_10588543
|
0.475
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr14_-_61191027
|
0.472
|
|
SIX4
|
SIX homeobox 4
|
|
chr18_+_32073245
|
0.471
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr8_-_6420454
|
0.471
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr7_+_129906702
|
0.466
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr11_+_92085261
|
0.465
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr5_-_132161863
|
0.462
|
NM_133456
|
SHROOM1
|
shroom family member 1
|
|
chr3_-_167371701
|
0.455
|
|
WDR49
|
WD repeat domain 49
|
|
chr4_+_88571453
|
0.445
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr5_-_39394423
|
0.439
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_+_32687246
|
0.437
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr1_-_27998658
|
0.432
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr2_+_210444154
|
0.429
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr15_-_60919642
|
0.416
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_+_89182038
|
0.414
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr6_-_76203256
|
0.412
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr15_+_78558522
|
0.407
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr6_+_30856462
|
0.407
|
NM_001202521
NM_001202522
NM_013994
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr4_+_89299874
|
0.403
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr18_+_32073343
|
0.401
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr6_-_56716713
|
0.399
|
|
DST
|
dystonin
|
|
chr1_-_27998719
|
0.386
|
NM_002038
NM_022872
NM_022873
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr12_-_71031174
|
0.384
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr1_-_33430285
|
0.381
|
NM_001127361
NM_153341
|
RNF19B
|
ring finger protein 19B
|
|
chr2_-_133539627
|
0.379
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr1_+_162602198
|
0.379
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr3_-_167371288
|
0.375
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr14_-_57960432
|
0.373
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr15_-_72563627
|
0.367
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr2_+_143635225
|
0.365
|
|
KYNU
|
kynureninase
|
|
chr4_+_108814419
|
0.363
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr15_+_42696957
|
0.362
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr11_-_18258315
|
0.362
|
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr13_+_97794050
|
0.361
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr6_-_56507585
|
0.360
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr16_-_30582951
|
0.356
|
|
ZNF688
|
zinc finger protein 688
|
|
chr12_+_7053180
|
0.355
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr14_-_23445885
|
0.352
|
|
AJUBA
|
ajuba LIM protein
|
|
chr11_-_10590078
|
0.347
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr10_-_74283693
|
0.346
|
NM_001195519
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr6_+_21666645
|
0.339
|
|
LINC00340
|
long intergenic non-protein coding RNA 340
|
|
chrX_-_108725284
|
0.337
|
NM_001522
|
GUCY2F
|
guanylate cyclase 2F, retinal
|
|
chr1_+_157963062
|
0.335
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr2_+_210444364
|
0.333
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr17_-_39092714
|
0.332
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr7_-_16921596
|
0.330
|
NM_176813
|
AGR3
|
anterior gradient 3 homolog (Xenopus laevis)
|
|
chr11_+_13984139
|
0.329
|
NM_006108
|
SPON1
|
spondin 1, extracellular matrix protein
|
|
chr1_-_178840079
|
0.329
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr11_+_46694514
|
0.328
|
|
ATG13
|
ATG13 autophagy related 13 homolog (S. cerevisiae)
|
|
chr7_+_28475233
|
0.327
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_+_143635194
|
0.326
|
NM_001032998
NM_001199241
NM_003937
|
KYNU
|
kynureninase
|
|
chr17_+_7384720
|
0.326
|
NM_001102614
|
SLC35G6
|
solute carrier family 35, member G6
|
|
chr10_+_96698409
|
0.325
|
NM_000771
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9
|
|
chr10_-_31288420
|
0.324
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr11_+_112047088
|
0.324
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr10_-_92680784
|
0.324
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr16_+_89984286
|
0.322
|
NM_002386
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor)
|
|
chr3_-_114102488
|
0.321
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_-_79212884
|
0.321
|
NM_144679
|
C17orf56
|
chromosome 17 open reading frame 56
|
|
chr3_-_87325611
|
0.320
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr10_-_32667543
|
0.316
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr5_-_39425030
|
0.312
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr1_+_145524989
|
0.302
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr2_+_169659099
|
0.301
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr1_-_57431620
|
0.295
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr5_+_137673223
|
0.294
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr6_+_32940860
|
0.293
|
NM_001199456
|
BRD2
|
bromodomain containing 2
|
|
chr13_-_52980628
|
0.292
|
NM_018676
NM_199263
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr7_+_112063131
|
0.290
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr20_-_62587718
|
0.290
|
NM_017859
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr14_-_92198383
|
0.287
|
NM_024764
|
CATSPERB
|
cation channel, sperm-associated, beta
|
|
chr15_-_77154284
|
0.285
|
NM_001145923
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr4_-_153303663
|
0.285
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr17_-_15523017
|
0.281
|
NM_006382
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr12_-_15865847
|
0.281
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr6_+_26104103
|
0.280
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chr5_+_68800003
|
0.280
|
NM_001205255
|
OCLN
|
occludin
|
|
chr4_+_54292035
|
0.277
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr10_-_11574273
|
0.277
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr22_-_39636906
|
0.277
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr17_-_79212775
|
0.275
|
|
C17orf56
|
chromosome 17 open reading frame 56
|
|
chr11_+_124481451
|
0.272
|
NM_052959
|
PANX3
|
pannexin 3
|
|
chr9_-_85882093
|
0.272
|
NM_001244962
|
FRMD3
|
FERM domain containing 3
|
|
chr6_+_24357130
|
0.270
|
NM_181337
|
KAAG1
|
kidney associated antigen 1
|
|
chr4_-_82393025
|
0.270
|
NM_152545
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr5_+_71502700
|
0.269
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr12_-_10573041
|
0.269
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr2_-_69098502
|
0.269
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chrX_-_76814217
|
0.268
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr3_+_136649316
|
0.268
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr11_+_73019662
|
0.267
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr18_+_55816546
|
0.261
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr11_-_59633942
|
0.259
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr19_+_36239261
|
0.258
|
NM_019104
|
LIN37
|
lin-37 homolog (C. elegans)
|
|
chr21_+_33664123
|
0.256
|
NM_178817
NM_206898
|
MRAP
|
melanocortin 2 receptor accessory protein
|
|
chr10_+_5936348
|
0.255
|
NM_032807
|
FBXO18
|
F-box protein, helicase, 18
|
|
chr5_+_172484353
|
0.253
|
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr5_-_145483877
|
0.252
|
NM_001029869
|
PLAC8L1
|
PLAC8-like 1
|
|
chr3_-_57530067
|
0.250
|
NM_178504
NM_198564
|
DNAH12
|
dynein, axonemal, heavy chain 12
|
|
chr15_+_74907334
|
0.250
|
NM_001130028
|
CLK3
|
CDC-like kinase 3
|
|
chr1_-_27998700
|
0.246
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr3_+_63953419
|
0.244
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr5_-_139937677
|
0.243
|
NM_001035235
|
SRA1
|
steroid receptor RNA activator 1
|
|
chr1_-_155774769
|
0.242
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr16_-_21436657
|
0.241
|
NM_130464
|
NPIPL3
|
nuclear pore complex interacting protein-like 3
|
|
chr2_-_161130151
|
0.240
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr5_+_149865310
|
0.239
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr15_-_69707358
|
0.237
|
|
LOC145694
|
uncharacterized LOC145694
|
|
chr22_+_30752612
|
0.236
|
NM_001017437
|
CCDC157
|
coiled-coil domain containing 157
|
|
chr1_-_179112178
|
0.236
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr3_+_38032260
|
0.235
|
|
VILL
|
villin-like
|
|
chr1_+_162602308
|
0.232
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr3_-_99833348
|
0.231
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr10_-_52645430
|
0.229
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr6_-_152702329
|
0.228
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr16_+_27341305
|
0.226
|
|
IL4R
|
interleukin 4 receptor
|
|
chr2_-_170430312
|
0.224
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr5_+_55149149
|
0.222
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr6_-_152702450
|
0.222
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr14_+_39583426
|
0.220
|
NM_001009182
NM_001009183
NM_003616
|
GEMIN2
|
gem (nuclear organelle) associated protein 2
|
|
chr5_-_159846043
|
0.219
|
NM_006425
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae)
|
|
chr14_+_105267378
|
0.218
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr19_+_13906207
|
0.217
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr10_+_94594469
|
0.215
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr2_-_145274916
|
0.214
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr9_+_139702395
|
0.212
|
|
C9orf86
|
chromosome 9 open reading frame 86
|
|
chr12_+_50869531
|
0.211
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr18_+_29769986
|
0.211
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr3_+_187943192
|
0.210
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_-_39195101
|
0.209
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr19_-_4338750
|
0.208
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr6_-_152623213
|
0.205
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr11_-_18258383
|
0.205
|
NM_006512
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr3_-_187454247
|
0.204
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_-_211018294
|
0.203
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr9_-_86432546
|
0.202
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr3_-_12586962
|
0.202
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr10_-_124605690
|
0.201
|
NM_022034
|
CUZD1
|
CUB and zona pellucida-like domains 1
|
|
chr9_-_3223007
|
0.201
|
|
|
|
|
chr4_+_74286846
|
0.200
|
|
ALB
|
albumin
|
|
chr11_+_124439892
|
0.199
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr16_-_52581713
|
0.198
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr9_+_139702403
|
0.197
|
|
C9orf86
|
chromosome 9 open reading frame 86
|
|
chr3_-_187452669
|
0.194
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr13_+_24844846
|
0.194
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr9_+_139702355
|
0.193
|
NM_001173988
NM_001173989
NM_024718
|
C9orf86
|
chromosome 9 open reading frame 86
|
|
chr17_-_29641094
|
0.193
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr7_-_141673572
|
0.189
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr17_-_38978824
|
0.189
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr3_-_114790221
|
0.189
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr19_+_48758931
|
0.189
|
|
LOC100505812
|
uncharacterized LOC100505812
|
|
chr11_-_14386051
|
0.188
|
NM_001177314
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2
|
|
chr3_+_184529871
|
0.188
|
NM_001009921
NM_015303
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae)
|
|
chr6_+_26156558
|
0.188
|
NM_005321
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr14_-_21566728
|
0.187
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr3_+_138066489
|
0.187
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr8_-_42234562
|
0.187
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr1_+_68297985
|
0.186
|
|
LOC100289178
|
uncharacterized LOC100289178
|
|
chr10_+_86184685
|
0.185
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr9_-_5968367
|
0.185
|
|
|
|
|
chr2_-_100722328
|
0.183
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr5_-_137848517
|
0.182
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr2_-_134326030
|
0.182
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr12_-_109027565
|
0.180
|
|
SELPLG
|
selectin P ligand
|
|
chr6_-_33754712
|
0.178
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr2_-_206950895
|
0.177
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chrM_+_4770
|
0.177
|
|
|
|
|
chr9_+_103947316
|
0.176
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr7_-_83270746
|
0.175
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr19_-_15391261
|
0.175
|
NM_014299
NM_058243
|
BRD4
|
bromodomain containing 4
|
|
chr14_+_37126772
|
0.175
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr2_-_135782246
|
0.174
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr2_-_216242916
|
0.174
|
|
FN1
|
fibronectin 1
|
|
chr19_+_2232071
|
0.173
|
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|