|
chr7_+_18535351
|
6.568
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
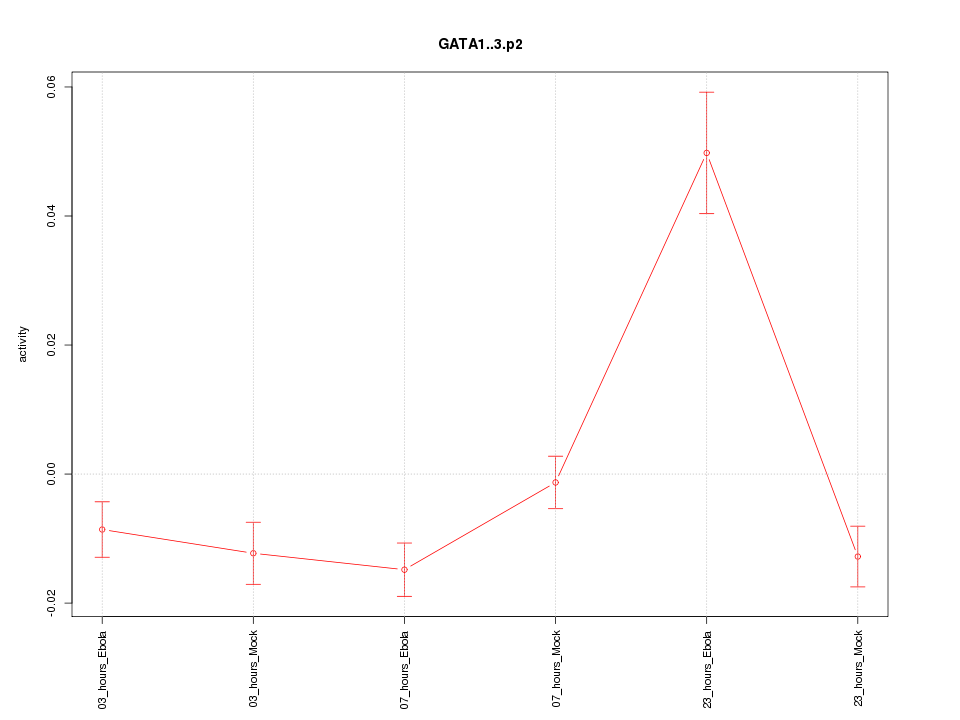
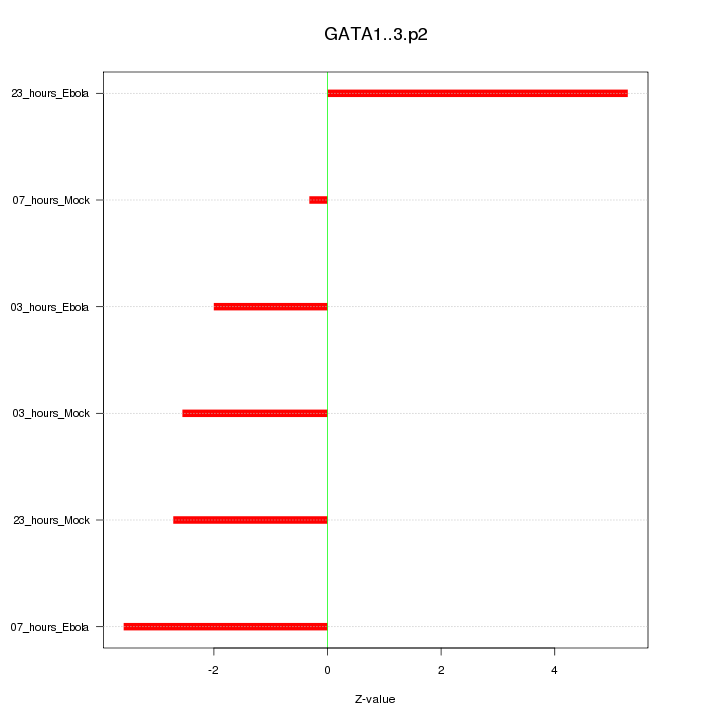
HDAC9
|
histone deacetylase 9
|
|
chr1_+_212788359
|
6.299
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr10_-_92681025
|
5.065
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr3_-_48700317
|
4.139
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr19_+_49376488
|
3.947
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr1_+_212738675
|
3.871
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr10_-_92680784
|
3.783
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr3_+_51989329
|
3.534
|
NM_080865
|
GPR62
|
G protein-coupled receptor 62
|
|
chr14_-_61191027
|
3.358
|
|
SIX4
|
SIX homeobox 4
|
|
chrX_+_70364680
|
3.268
|
NM_001166660
NM_018977
NM_181303
|
NLGN3
|
neuroligin 3
|
|
chr6_+_17282931
|
3.117
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_-_74734498
|
2.907
|
|
PCGF1
|
polycomb group ring finger 1
|
|
chr3_-_114343791
|
2.632
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr14_-_61190458
|
2.624
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr2_-_145277879
|
2.599
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_-_114343052
|
2.544
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_-_145274916
|
2.519
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_+_49126480
|
2.502
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr2_-_74734785
|
2.442
|
NM_032673
|
PCGF1
|
polycomb group ring finger 1
|
|
chr7_+_18535817
|
2.416
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr12_+_5603297
|
2.325
|
NM_002527
|
NTF3
|
neurotrophin 3
|
|
chr17_+_7743233
|
2.324
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr7_+_39017608
|
2.322
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chrX_+_95939661
|
2.303
|
NM_006729
NM_007309
|
DIAPH2
|
diaphanous homolog 2 (Drosophila)
|
|
chr4_+_75230856
|
2.299
|
NM_001432
|
EREG
|
epiregulin
|
|
chr5_-_148758787
|
2.249
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr11_+_124439892
|
2.217
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr14_+_55221505
|
2.202
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr2_-_69098502
|
2.176
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr17_-_72968750
|
2.162
|
|
|
|
|
chr9_-_3525721
|
2.129
|
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr20_-_44600939
|
2.086
|
|
ZNF335
|
zinc finger protein 335
|
|
chr11_+_5968576
|
2.085
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chrX_+_70364707
|
2.084
|
|
NLGN3
|
neuroligin 3
|
|
chr4_-_89618928
|
2.081
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr11_-_9225780
|
2.063
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr15_+_93443559
|
2.054
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr6_+_37137882
|
2.054
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr2_-_208994408
|
2.038
|
NM_020989
|
CRYGC
|
crystallin, gamma C
|
|
chr17_-_17399494
|
2.035
|
NM_001199989
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr3_-_49170427
|
2.015
|
|
LAMB2
|
laminin, beta 2 (laminin S)
|
|
chr15_+_93443418
|
2.012
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr15_+_71839540
|
1.988
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr3_-_49170490
|
1.947
|
NM_002292
|
LAMB2
|
laminin, beta 2 (laminin S)
|
|
chr3_-_114478117
|
1.915
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_51993955
|
1.885
|
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr19_-_50316360
|
1.865
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr1_+_35331533
|
1.852
|
|
|
|
|
chr6_-_32160289
|
1.851
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr4_-_184243578
|
1.818
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chrX_+_69642880
|
1.795
|
NM_001171191
NM_001171192
NM_001171193
NM_017711
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2
|
|
chr11_-_10590078
|
1.791
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr22_+_31478845
|
1.782
|
NM_001207017
|
SMTN
|
smoothelin
|
|
chr9_-_34397786
|
1.753
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr9_+_35560424
|
1.751
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr1_+_231298673
|
1.744
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr9_-_34662629
|
1.738
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr2_-_145277568
|
1.722
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr14_+_21538339
|
1.720
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr1_+_25598883
|
1.698
|
NM_001127691
NM_016124
|
RHD
|
Rh blood group, D antigen
|
|
chr1_+_178062829
|
1.695
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr10_-_77161424
|
1.693
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr11_-_83984230
|
1.693
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr10_-_77161136
|
1.686
|
|
ZNF503
|
zinc finger protein 503
|
|
chr4_+_118955499
|
1.684
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr1_+_26644380
|
1.675
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chrX_+_49126326
|
1.668
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr1_-_153930911
|
1.667
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chrX_+_95939851
|
1.666
|
|
DIAPH2
|
diaphanous homolog 2 (Drosophila)
|
|
chrX_+_69488154
|
1.664
|
NM_004312
|
ARR3
|
arrestin 3, retinal (X-arrestin)
|
|
chr1_-_145715561
|
1.658
|
NM_007053
|
CD160
|
CD160 molecule
|
|
chr12_-_10978867
|
1.657
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chr20_-_44600787
|
1.652
|
NM_022095
|
ZNF335
|
zinc finger protein 335
|
|
chr6_-_76203256
|
1.652
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr18_+_32398225
|
1.646
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr15_-_60919642
|
1.642
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chrX_+_49126299
|
1.640
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr16_+_69680959
|
1.626
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr11_-_47470687
|
1.626
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr15_+_41136622
|
1.615
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr7_-_27135524
|
1.609
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr20_-_31172874
|
1.609
|
|
LOC284804
|
uncharacterized LOC284804
|
|
chr19_+_13135809
|
1.605
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr7_-_100808832
|
1.604
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr5_+_137673956
|
1.590
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr12_-_15114495
|
1.574
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr19_-_46295647
|
1.570
|
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr2_+_149632772
|
1.561
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr19_+_13135745
|
1.552
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr15_+_78558522
|
1.536
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr19_+_8386374
|
1.535
|
NM_001031
|
RPS28
|
ribosomal protein S28
|
|
chr7_+_27135672
|
1.532
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr7_-_44105162
|
1.529
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr7_+_28725597
|
1.528
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_-_3525973
|
1.520
|
NM_002919
NM_134428
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr7_+_18330044
|
1.518
|
|
HDAC9
|
histone deacetylase 9
|
|
chr5_+_137673953
|
1.507
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr8_-_29207795
|
1.505
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr6_+_32939974
|
1.496
|
|
BRD2
|
bromodomain containing 2
|
|
chr16_+_67573746
|
1.480
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr19_+_13906207
|
1.474
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr1_+_27101217
|
1.461
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr7_+_23530136
|
1.451
|
|
RPS2P32
|
ribosomal protein S2 pseudogene 32
|
|
chr16_+_1756161
|
1.449
|
NM_001040439
NM_015133
|
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr4_-_83347778
|
1.446
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chrX_+_70521627
|
1.436
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr1_-_113161724
|
1.432
|
NM_017744
NM_138727
NM_138728
NM_138729
|
ST7L
|
suppression of tumorigenicity 7 like
|
|
chr1_+_161195832
|
1.429
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr4_+_88571453
|
1.424
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr17_+_7758380
|
1.419
|
NM_203411
|
TMEM88
|
transmembrane protein 88
|
|
chr20_+_61867234
|
1.409
|
NM_022161
NM_139317
|
BIRC7
|
baculoviral IAP repeat containing 7
|
|
chr1_-_149908759
|
1.402
|
NM_001145862
|
MTMR11
|
myotubularin related protein 11
|
|
chr14_-_23446431
|
1.396
|
NM_198086
|
AJUBA
|
ajuba LIM protein
|
|
chr6_-_90024936
|
1.384
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr22_-_39268169
|
1.380
|
|
CBX6
|
chromobox homolog 6
|
|
chr18_+_32335939
|
1.376
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr7_-_79082870
|
1.372
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr19_-_56056908
|
1.370
|
NM_001199824
|
SGK110
|
putative uncharacterized serine/threonine-protein kinase SgK110-like
|
|
chr8_-_42698447
|
1.362
|
NM_018105
NM_199003
|
THAP1
|
THAP domain containing, apoptosis associated protein 1
|
|
chr5_+_137673702
|
1.362
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr19_+_55795533
|
1.359
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr19_+_54376975
|
1.353
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr20_-_1309782
|
1.343
|
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr7_+_64498643
|
1.339
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr2_+_149633173
|
1.336
|
|
|
|
|
chr17_-_72968790
|
1.335
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr12_-_48597074
|
1.335
|
NM_001004134
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1
|
|
chr10_+_74451838
|
1.332
|
NM_138357
|
MCU
|
mitochondrial calcium uniporter
|
|
chrM_+_10123
|
1.332
|
|
|
|
|
chr19_-_3969734
|
1.332
|
NM_001348
|
DAPK3
|
death-associated protein kinase 3
|
|
chr3_-_126194710
|
1.331
|
|
ZXDC
|
ZXD family zinc finger C
|
|
chr7_-_140302311
|
1.319
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr16_-_29937354
|
1.317
|
|
KCTD13
|
potassium channel tetramerisation domain containing 13
|
|
chr2_+_149632836
|
1.314
|
|
KIF5C
|
kinesin family member 5C
|
|
chr6_+_31515352
|
1.313
|
NM_001144961
NM_005007
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr4_+_74606222
|
1.304
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr5_-_172198160
|
1.303
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr1_+_109242188
|
1.281
|
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr6_+_149721494
|
1.280
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr6_+_37252166
|
1.278
|
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr8_-_124665165
|
1.278
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr11_+_67034164
|
1.278
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr3_-_169899519
|
1.270
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr14_-_23445885
|
1.267
|
|
AJUBA
|
ajuba LIM protein
|
|
chr3_-_172166202
|
1.262
|
NM_004122
NM_198407
|
GHSR
|
growth hormone secretagogue receptor
|
|
chr1_-_150780705
|
1.262
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr20_-_44516237
|
1.261
|
NM_080608
|
SPATA25
|
spermatogenesis associated 25
|
|
chr11_+_65984213
|
1.254
|
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr17_+_19314490
|
1.246
|
NM_007148
|
RNF112
|
ring finger protein 112
|
|
chr6_-_44233244
|
1.235
|
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr6_-_53013581
|
1.232
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr6_-_30655078
|
1.231
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr8_-_42234562
|
1.227
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr7_+_128312345
|
1.225
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr13_+_42712177
|
1.225
|
NM_001204505
NM_001204506
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr1_-_204183036
|
1.220
|
NM_198447
|
GOLT1A
|
golgi transport 1A
|
|
chr14_-_70263871
|
1.215
|
NM_003049
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1
|
|
chr3_-_126194761
|
1.203
|
NM_001040653
NM_025112
|
ZXDC
|
ZXD family zinc finger C
|
|
chr4_-_88243994
|
1.202
|
NM_001136230
NM_178135
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13
|
|
chr9_-_35618300
|
1.194
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr1_+_45266048
|
1.191
|
|
PLK3
|
polo-like kinase 3
|
|
chr3_+_119421865
|
1.189
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr3_+_19189969
|
1.188
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr22_-_39268214
|
1.176
|
|
CBX6
|
chromobox homolog 6
|
|
chr19_-_46272496
|
1.175
|
NM_175875
|
SIX5
|
SIX homeobox 5
|
|
chr19_+_41281281
|
1.175
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr17_-_39092714
|
1.168
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_+_26020677
|
1.167
|
NM_003529
|
HIST1H3A
|
histone cluster 1, H3a
|
|
chr10_-_75335415
|
1.166
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr12_+_50024326
|
1.165
|
NM_001031698
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr19_-_1848359
|
1.163
|
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)
|
|
chr19_+_56124958
|
1.157
|
NM_001195605
|
ZNF865
|
zinc finger protein 865
|
|
chr1_-_120484230
|
1.156
|
|
NOTCH2
|
notch 2
|
|
chr22_+_51039178
|
1.152
|
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr12_-_99548471
|
1.151
|
NM_001204068
NM_001204069
NM_001204070
NM_020140
NM_181670
NM_001204066
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chrM_+_1670
|
1.145
|
|
|
|
|
chr11_+_61447816
|
1.145
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr17_-_7106561
|
1.144
|
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr6_-_11382501
|
1.139
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_+_37571752
|
1.137
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr2_+_210444364
|
1.121
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_-_201915467
|
1.119
|
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr6_+_90276556
|
1.118
|
NM_001242814
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr4_-_114900802
|
1.118
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr10_-_98945662
|
1.115
|
NM_003061
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr17_-_73774447
|
1.111
|
|
|
|
|
chr11_+_67034148
|
1.108
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr10_-_32667543
|
1.102
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr9_-_4152182
|
1.101
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_-_208030613
|
1.100
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr17_+_41003184
|
1.099
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr15_-_60884615
|
1.099
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_49591897
|
1.094
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr1_-_193155723
|
1.091
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr4_+_74286846
|
1.090
|
|
ALB
|
albumin
|
|
chr5_+_137801178
|
1.087
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr2_-_145275086
|
1.087
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_-_59248287
|
1.084
|
|
JUN
|
jun proto-oncogene
|
|
chr2_+_74273449
|
1.077
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr4_+_123161119
|
1.074
|
|
KIAA1109
|
KIAA1109
|