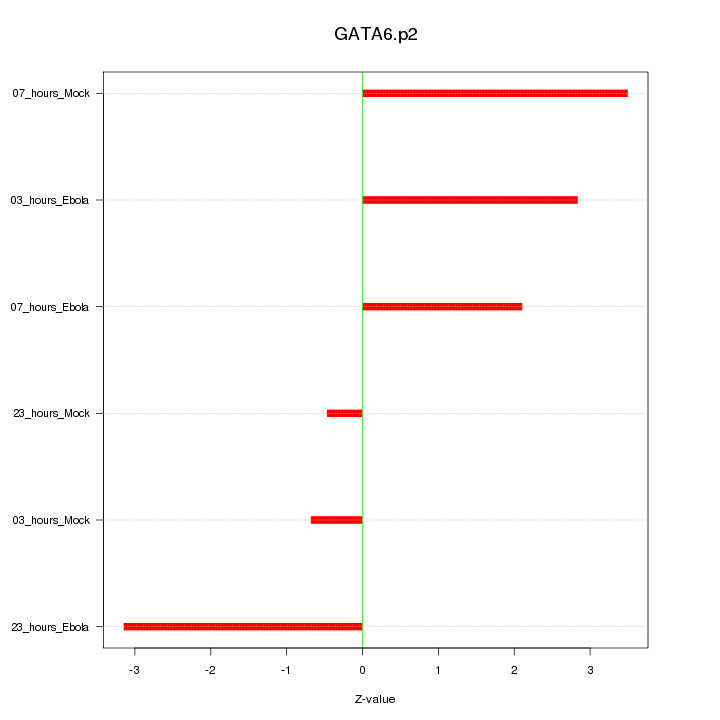
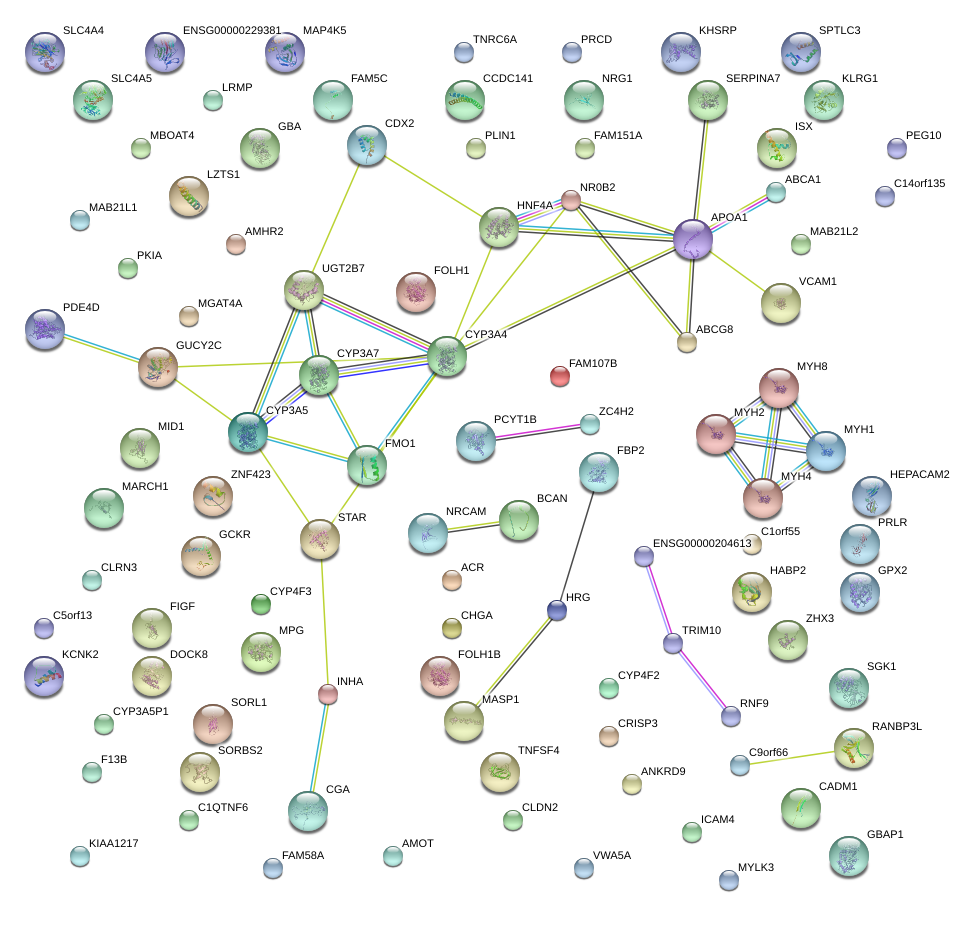
|
chr5_-_36301999
|
4.666
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr9_-_107665959
|
4.066
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr9_-_107666046
|
3.943
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr12_+_25205180
|
3.658
|
NM_001204126
NM_001204127
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chrX_-_105282714
|
3.470
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr11_-_115088628
|
3.458
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr3_+_186383770
|
2.889
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr10_-_129691202
|
2.704
|
NM_152311
|
CLRN3
|
clarin 3
|
|
chr13_-_28543143
|
2.558
|
|
CDX2
|
caudal type homeobox 2
|
|
chr10_-_129691047
|
2.479
|
|
|
|
|
chr2_-_179914785
|
2.473
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr6_-_30128688
|
2.467
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr22_+_35462125
|
2.355
|
NM_001008494
|
ISX
|
intestine-specific homeobox
|
|
chr13_-_28543423
|
2.275
|
|
CDX2
|
caudal type homeobox 2
|
|
chr8_+_79428335
|
2.223
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr8_-_30002199
|
2.159
|
NM_001100916
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4
|
|
chr6_-_49712055
|
2.028
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr12_+_25205488
|
1.930
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_+_7169192
|
1.862
|
|
|
|
|
chr20_+_12989616
|
1.852
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr20_+_43029973
|
1.841
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr19_+_10397642
|
1.672
|
NM_001039132
NM_001544
NM_022377
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|
|
chr11_+_121461070
|
1.656
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr6_-_134498973
|
1.641
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chrX_+_106161589
|
1.611
|
NM_001171095
|
CLDN2
|
claudin 2
|
|
chr8_-_38008332
|
1.569
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chrX_+_106163605
|
1.561
|
NM_020384
|
CLDN2
|
claudin 2
|
|
chr1_+_171217632
|
1.486
|
NM_002021
|
FMO1
|
flavin containing monooxygenase 1
|
|
chr4_+_151503076
|
1.460
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr13_-_28543290
|
1.438
|
NM_001265
|
CDX2
|
caudal type homeobox 2
|
|
chr4_+_69962191
|
1.435
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr1_-_173176314
|
1.427
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chrX_-_15402469
|
1.408
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr16_-_46782104
|
1.388
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr20_+_43029865
|
1.379
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr7_-_99332715
|
1.374
|
NM_000765
|
CYP3A7
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 7
cytochrome P450, family 3, subfamily A, polypeptide 5
|
|
chr1_+_215178884
|
1.373
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr1_-_155211049
|
1.344
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chrX_-_112066353
|
1.340
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr20_-_39928708
|
1.275
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr11_+_89392464
|
1.259
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr19_+_15751706
|
1.235
|
NM_000896
NM_001199208
NM_001199209
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr4_-_164534656
|
1.225
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr2_+_44066102
|
1.223
|
NM_022437
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr15_-_90222572
|
1.219
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr2_+_27719702
|
1.218
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chrX_-_64196268
|
1.210
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr2_-_74570533
|
1.198
|
NM_133478
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chrX_-_24690740
|
1.196
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr1_-_190446701
|
1.185
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr9_-_215892
|
1.176
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr6_-_87804773
|
1.132
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr2_+_220436953
|
1.125
|
NM_002191
|
INHA
|
inhibin, alpha
|
|
chr5_-_35118223
|
1.117
|
NM_001204314
NM_001204315
NM_001204316
NM_001204317
NM_001204318
|
PRLR
|
prolactin receptor
|
|
chr16_+_24802421
|
1.105
|
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr8_-_20112691
|
1.080
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr16_-_49860917
|
1.070
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr5_-_111312522
|
1.069
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_27240430
|
1.059
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr14_-_65409445
|
1.058
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr9_+_273025
|
1.054
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr2_-_99279935
|
1.040
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr11_-_116708334
|
1.037
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr12_+_9142136
|
1.027
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr17_+_74536114
|
0.988
|
NM_001077620
|
PRCD
|
progressive rod-cone degeneration
|
|
chr3_-_187009469
|
0.984
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr9_-_97356007
|
0.975
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chr1_+_101185195
|
0.953
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr11_+_123986103
|
0.950
|
NM_001130142
NM_014622
NM_198315
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr7_+_94292737
|
0.933
|
|
PEG10
|
paternally expressed 10
|
|
chr10_-_14816895
|
0.927
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr10_+_24755440
|
0.913
|
|
KIAA1217
|
KIAA1217
|
|
chr1_-_197036350
|
0.909
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr10_+_115310589
|
0.906
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr5_-_59783885
|
0.897
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_226187019
|
0.894
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chrX_-_10851808
|
0.893
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr7_-_92855780
|
0.892
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr1_-_55089068
|
0.888
|
NM_176782
|
FAM151A
|
family with sequence similarity 151, member A
|
|
chr17_-_10372875
|
0.876
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr12_+_53818864
|
0.870
|
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr4_-_186697065
|
0.861
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr22_+_51176623
|
0.850
|
NM_001097
|
ACR
|
acrosin
|
|
chr22_-_37584254
|
0.844
|
NM_031910
NM_182486
|
C1QTNF6
|
C1q and tumor necrosis factor related protein 6
|
|
chr7_-_107880492
|
0.841
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr4_-_159094163
|
0.826
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_140557370
|
0.822
|
NM_019120
|
PCDHB8
|
protocadherin beta 8
|
|
chr10_+_75673346
|
0.822
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr6_-_52668599
|
0.817
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr12_-_23737507
|
0.813
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_+_39408541
|
0.811
|
|
KLB
|
klotho beta
|
|
chrX_-_24665225
|
0.811
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr12_+_123944385
|
0.800
|
NM_180699
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12)
|
|
chr15_-_50838901
|
0.791
|
NM_203494
|
USP50
|
ubiquitin specific peptidase 50
|
|
chr10_+_24755459
|
0.790
|
|
KIAA1217
|
KIAA1217
|
|
chr7_+_73245192
|
0.786
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr18_-_21891469
|
0.786
|
NM_001242508
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr8_-_95449131
|
0.769
|
NM_001205263
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chr17_-_56084571
|
0.762
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr17_-_56084590
|
0.759
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr1_-_232651227
|
0.758
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr7_+_95115212
|
0.748
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr15_-_76304762
|
0.738
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr7_-_150945731
|
0.728
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr19_-_36303766
|
0.710
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr3_+_49568370
|
0.702
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr20_-_45976626
|
0.701
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr6_-_2840683
|
0.696
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr1_+_81771844
|
0.696
|
|
LPHN2
|
latrophilin 2
|
|
chr5_+_140602914
|
0.693
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr15_-_55652777
|
0.692
|
|
CCPG1
|
cell cycle progression 1
|
|
chr4_+_120056938
|
0.680
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr1_-_226187027
|
0.672
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr3_+_100328432
|
0.669
|
NM_032787
|
GPR128
|
G protein-coupled receptor 128
|
|
chr5_+_140579182
|
0.667
|
NM_018931
|
PCDHB11
|
protocadherin beta 11
|
|
chr1_-_217113007
|
0.664
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr18_-_64271215
|
0.662
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr6_+_130339715
|
0.656
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr3_-_15563160
|
0.648
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr1_-_31196429
|
0.645
|
NM_002379
|
MATN1
|
matrilin 1, cartilage matrix protein
|
|
chr2_+_96991934
|
0.644
|
NM_178495
NM_001163524
|
ITPRIPL1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1
|
|
chr20_+_54967692
|
0.641
|
NM_001033522
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa
|
|
chr7_+_128379428
|
0.641
|
|
CALU
|
calumenin
|
|
chr18_+_29171729
|
0.640
|
NM_000371
|
TTR
|
transthyretin
|
|
chr7_+_128379400
|
0.637
|
|
CALU
|
calumenin
|
|
chr1_-_120311466
|
0.631
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr12_-_57410275
|
0.625
|
NM_001178054
NM_013251
|
TAC3
|
tachykinin 3
|
|
chr7_+_128379452
|
0.624
|
|
CALU
|
calumenin
|
|
chr13_-_46756295
|
0.622
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr2_-_88427535
|
0.620
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chrX_-_10535476
|
0.610
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr3_-_119379269
|
0.608
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr4_-_76957139
|
0.602
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr1_+_63063186
|
0.597
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr4_+_110834039
|
0.596
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr3_-_186262114
|
0.594
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|
|
chr9_+_118916069
|
0.589
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr2_+_211458078
|
0.586
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr4_-_70518964
|
0.584
|
NM_001252274
NM_001252275
NM_006798
|
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus
|
|
chr3_-_52488030
|
0.580
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chrX_+_99899162
|
0.577
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr5_-_154230212
|
0.573
|
NM_032385
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr6_+_26365397
|
0.568
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr3_-_112359863
|
0.567
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chrX_-_100307042
|
0.566
|
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr11_+_61248584
|
0.563
|
NM_001170753
NM_145017
|
PPP1R32
|
protein phosphatase 1, regulatory subunit 32
|
|
chr17_+_7517263
|
0.559
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr1_-_226186987
|
0.558
|
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chrX_-_55020510
|
0.556
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr11_+_72983246
|
0.555
|
NM_004154
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr13_-_49975734
|
0.554
|
NM_030925
|
CAB39L
|
calcium binding protein 39-like
|
|
chr16_+_24788568
|
0.554
|
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr16_+_24857551
|
0.547
|
NM_052944
|
SLC5A11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11
|
|
chr12_-_70004941
|
0.543
|
NM_201550
|
LRRC10
|
leucine rich repeat containing 10
|
|
chr1_-_24469571
|
0.543
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr4_-_186733372
|
0.540
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_226187055
|
0.531
|
NM_152608
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr1_-_33647626
|
0.527
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chr1_-_149858114
|
0.527
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr12_+_86268072
|
0.525
|
NM_006183
|
NTS
|
neurotensin
|
|
chr22_+_21128378
|
0.525
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr3_-_168864325
|
0.524
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr5_+_42549657
|
0.521
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr22_+_35779103
|
0.519
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr12_+_56075329
|
0.517
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr7_-_150945701
|
0.508
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr6_-_13408368
|
0.506
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr1_+_244214576
|
0.500
|
|
ZNF238
|
zinc finger protein 238
|
|
chr1_-_39347258
|
0.498
|
NM_030772
|
GJA9-MYCBP
GJA9
|
GJA9-MYCBP readthrough
gap junction protein, alpha 9, 59kDa
|
|
chr3_+_195943382
|
0.497
|
NM_152672
|
OSTalpha
|
organic solute transporter alpha
|
|
chr19_+_20959109
|
0.491
|
|
|
|
|
chr7_+_129932973
|
0.486
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr13_-_46756326
|
0.486
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr12_+_53645369
|
0.483
|
NM_001170790
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr2_+_191301020
|
0.482
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr14_+_51026742
|
0.474
|
NM_015915
NM_181598
|
ATL1
|
atlastin GTPase 1
|
|
chr5_+_140501399
|
0.472
|
|
PCDHB4
|
protocadherin beta 4
|
|
chr9_+_684193
|
0.469
|
|
|
|
|
chr6_+_26402464
|
0.468
|
NM_001145008
NM_001145009
NM_007048
NM_194441
|
BTN3A1
|
butyrophilin, subfamily 3, member A1
|
|
chr17_-_72869068
|
0.468
|
NM_004110
NM_024417
|
FDXR
|
ferredoxin reductase
|
|
chr20_+_54967405
|
0.462
|
NM_001033521
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa
|
|
chr3_-_98312384
|
0.462
|
NM_000097
|
CPOX
|
coproporphyrinogen oxidase
|
|
chr17_+_43224683
|
0.459
|
NM_006460
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr6_-_117996369
|
0.458
|
|
|
|
|
chr13_-_46756291
|
0.456
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr20_+_54967572
|
0.452
|
NM_001324
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa
|
|
chr11_+_118958696
|
0.452
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr9_-_75567915
|
0.448
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr5_+_42565963
|
0.448
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chrX_+_22050920
|
0.446
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr17_+_6939236
|
0.444
|
NM_201566
|
SLC16A13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13)
|
|
chr3_+_155860750
|
0.443
|
NM_003471
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr1_+_50574601
|
0.441
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_-_168864066
|
0.435
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr6_-_31514620
|
0.434
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr3_-_112359565
|
0.430
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr19_-_13213513
|
0.429
|
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr2_-_165551178
|
0.427
|
|
COBLL1
|
COBL-like 1
|
|
chr21_-_36421195
|
0.425
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_57234279
|
0.424
|
NM_003865
|
HESX1
|
HESX homeobox 1
|