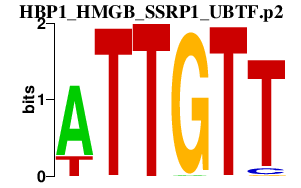
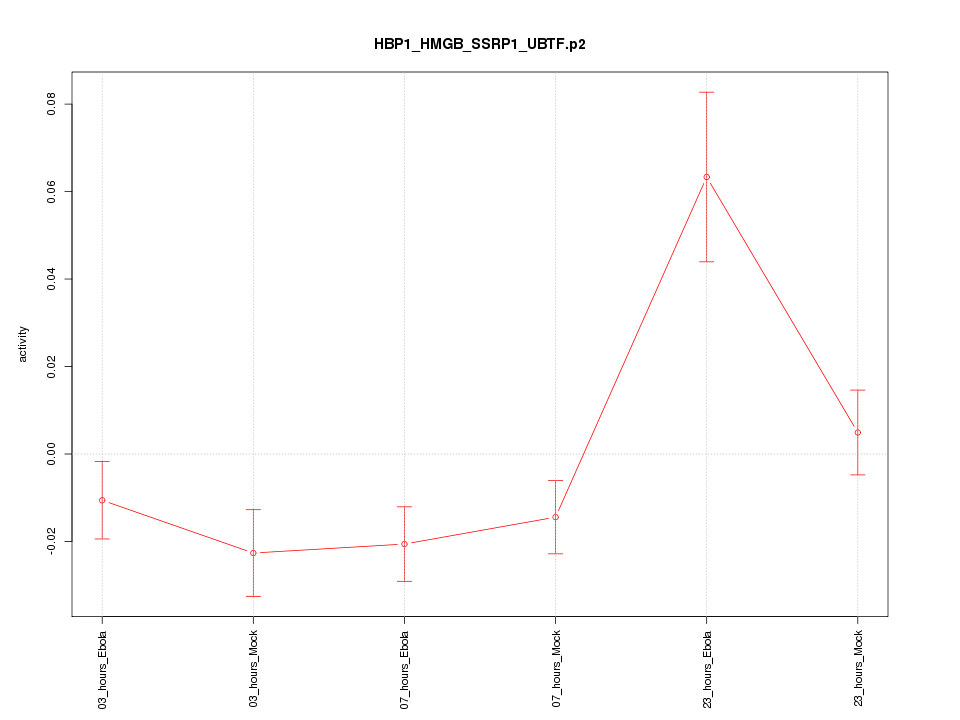
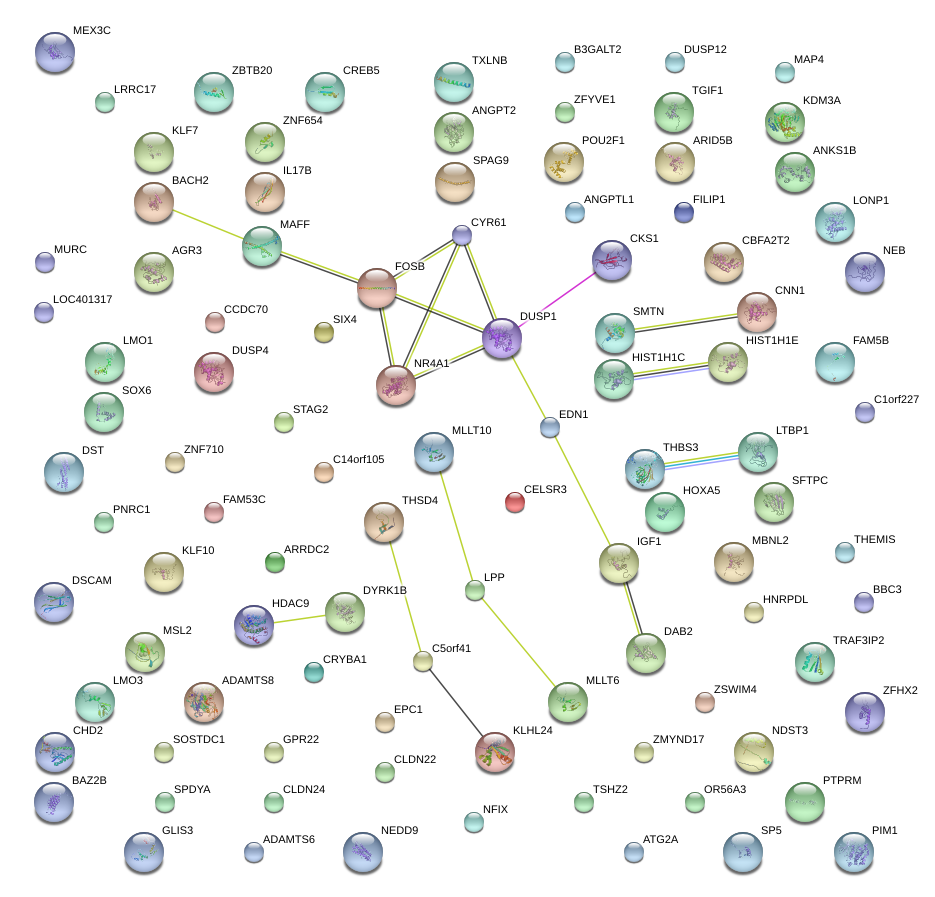
|
chr19_+_45971249
|
6.014
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr6_+_89790464
|
4.898
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr6_+_89790412
|
4.835
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr1_-_193155723
|
4.675
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr7_+_18535351
|
4.659
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr3_-_114790221
|
4.093
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr19_+_13134782
|
4.088
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr10_-_32667543
|
3.765
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr7_+_28452143
|
3.647
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_-_138189313
|
3.615
|
|
|
|
|
chr1_+_86046434
|
3.605
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr7_+_28725597
|
3.406
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr19_+_13906207
|
3.387
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr6_-_56716713
|
3.319
|
|
DST
|
dystonin
|
|
chrX_+_123234462
|
3.003
|
|
STAG2
|
stromal antigen 2
|
|
chr22_+_38598065
|
2.981
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr17_+_27573874
|
2.966
|
NM_005208
|
CRYBA1
|
crystallin, beta A1
|
|
chr19_-_47734215
|
2.921
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr8_-_6420454
|
2.907
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr4_-_184243578
|
2.878
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr12_-_16757944
|
2.866
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_-_11232901
|
2.835
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr3_-_47933997
|
2.791
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr6_-_11232883
|
2.757
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr11_-_16424391
|
2.738
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr17_+_36861398
|
2.669
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr11_+_5968576
|
2.595
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr3_-_114478117
|
2.588
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr22_+_38599026
|
2.524
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr4_-_83347778
|
2.513
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr6_-_11232900
|
2.505
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_76203256
|
2.459
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr5_+_172483285
|
2.453
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr2_-_152590983
|
2.329
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr9_-_4299915
|
2.321
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_+_149633173
|
2.313
|
|
|
|
|
chr6_+_12290528
|
2.304
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr12_+_52445185
|
2.304
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr3_-_48700317
|
2.299
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr7_-_16505292
|
2.295
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr18_+_3451589
|
2.274
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr6_-_91006460
|
2.266
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr7_+_28448970
|
2.264
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr6_-_91006580
|
2.254
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr4_+_118955499
|
2.253
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr5_+_137673223
|
2.232
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr6_+_21666645
|
2.163
|
|
LINC00340
|
long intergenic non-protein coding RNA 340
|
|
chr7_+_107110501
|
2.149
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr7_-_27183225
|
2.146
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr17_-_49124128
|
2.144
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr18_+_7946885
|
2.137
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr3_-_114866090
|
2.127
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_-_29206321
|
2.123
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr6_-_56716412
|
2.102
|
|
DST
|
dystonin
|
|
chr5_-_39425297
|
2.092
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr15_+_93443418
|
2.085
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr5_-_39424930
|
2.046
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr6_-_111888520
|
2.026
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr15_+_93443559
|
1.999
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr19_+_11649578
|
1.995
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr5_-_64777703
|
1.941
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr3_+_141105283
|
1.933
|
|
|
|
|
chr20_+_51588945
|
1.931
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr2_+_171571800
|
1.928
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr19_+_13135368
|
1.913
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_+_13135809
|
1.892
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr1_+_177140062
|
1.851
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr19_+_13135745
|
1.835
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr14_-_24020766
|
1.829
|
NM_033400
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr7_+_18535817
|
1.828
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr3_+_88188261
|
1.768
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr3_-_114343052
|
1.766
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_99548471
|
1.752
|
NM_001204068
NM_001204069
NM_001204070
NM_020140
NM_181670
NM_001204066
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr14_-_57960432
|
1.739
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr6_+_26156558
|
1.726
|
NM_005321
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr2_-_160472960
|
1.701
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr13_+_97794050
|
1.693
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr2_+_29038862
|
1.664
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr15_+_71433787
|
1.659
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr15_+_90544608
|
1.655
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr10_+_63661012
|
1.647
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr18_-_48724031
|
1.646
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr6_+_37137882
|
1.631
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr20_+_32150139
|
1.631
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr17_+_36861857
|
1.628
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr2_+_33359607
|
1.617
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr6_-_128222175
|
1.599
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr4_-_184241926
|
1.594
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr2_+_86668488
|
1.575
|
NM_001146688
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr14_-_73493744
|
1.573
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr3_-_135913276
|
1.556
|
NM_001145417
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr12_-_99548867
|
1.548
|
NM_001204067
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_+_103340335
|
1.543
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr2_+_86668428
|
1.539
|
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr13_-_60325073
|
1.536
|
|
|
|
|
chr19_-_40324747
|
1.528
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr7_+_18548899
|
1.528
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr3_+_187943192
|
1.524
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr15_+_71839540
|
1.518
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr1_-_155177685
|
1.514
|
NM_001252607
NM_001252608
NM_007112
|
THBS3
|
thrombospondin 3
|
|
chr1_+_167298280
|
1.509
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr19_+_18118921
|
1.494
|
NM_015683
|
ARRDC2
|
arrestin domain containing 2
|
|
chr22_+_31477231
|
1.491
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr10_-_75193297
|
1.490
|
NM_001024593
|
ZMYND17
|
zinc finger, MYND-type containing 17
|
|
chr3_+_183353355
|
1.483
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr2_+_86668270
|
1.470
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr7_-_16921596
|
1.463
|
NM_176813
|
AGR3
|
anterior gradient 3 homolog (Xenopus laevis)
|
|
chr8_-_103666018
|
1.445
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr5_-_172198160
|
1.442
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr13_+_97928414
|
1.436
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_+_177140530
|
1.431
|
NM_021165
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr6_-_139613114
|
1.429
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr1_-_213020947
|
1.415
|
NM_001024601
|
C1orf227
|
chromosome 1 open reading frame 227
|
|
chr5_-_39425030
|
1.411
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_-_102872329
|
1.395
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_178840079
|
1.395
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr14_-_61190458
|
1.385
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr7_+_102553343
|
1.378
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr13_+_52436116
|
1.371
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr6_+_26156552
|
1.370
|
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr2_-_208030613
|
1.365
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr5_-_148758787
|
1.363
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr11_-_64684606
|
1.356
|
NM_015104
|
ATG2A
|
ATG2 autophagy related 2 homolog A (S. cerevisiae)
|
|
chr2_+_169923531
|
1.355
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr3_-_187454247
|
1.347
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr1_+_203020310
|
1.346
|
NM_015053
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
|
|
chr6_+_7579692
|
1.346
|
|
DSP
|
desmoplakin
|
|
chr11_+_46299585
|
1.344
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr3_+_19189969
|
1.343
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr14_-_71275732
|
1.333
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr12_-_28124915
|
1.332
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr12_+_14538232
|
1.320
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr19_+_7660698
|
1.318
|
NM_001080429
NM_020902
|
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3
|
|
chr11_-_129062092
|
1.312
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr15_+_80364902
|
1.312
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr15_+_99191671
|
1.310
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr11_-_16497917
|
1.307
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr15_-_56209305
|
1.300
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_+_173472692
|
1.299
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr10_-_62332666
|
1.298
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr4_+_71494460
|
1.297
|
NM_031889
|
ENAM
|
enamelin
|
|
chr10_-_94003002
|
1.286
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr2_-_158182154
|
1.282
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr16_+_69726751
|
1.281
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr5_+_55147206
|
1.272
|
NM_001242637
NM_001242638
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr7_-_139876718
|
1.260
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr20_-_22566092
|
1.259
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr2_-_225370687
|
1.253
|
|
CUL3
|
cullin 3
|
|
chr20_-_22564893
|
1.234
|
|
FOXA2
|
forkhead box A2
|
|
chr9_-_4300028
|
1.234
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr10_-_11574273
|
1.231
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr11_-_84634437
|
1.221
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr10_-_14372807
|
1.213
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr10_+_112631675
|
1.209
|
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr15_+_83477972
|
1.205
|
NM_001080435
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chr3_+_136676706
|
1.199
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr10_-_13749622
|
1.192
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr5_-_39394423
|
1.188
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr2_+_109204904
|
1.186
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr7_+_18330044
|
1.184
|
|
HDAC9
|
histone deacetylase 9
|
|
chr16_+_50300461
|
1.175
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr5_+_172484353
|
1.175
|
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr1_-_221915458
|
1.172
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr22_+_38597938
|
1.169
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr9_-_4152182
|
1.167
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr1_-_221915393
|
1.166
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr19_-_3979828
|
1.164
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr1_-_221910801
|
1.160
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr5_+_78532338
|
1.155
|
|
|
|
|
chr13_+_97928196
|
1.152
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr13_+_24144402
|
1.151
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr2_+_228678556
|
1.148
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr12_-_117319231
|
1.148
|
NM_003806
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain)
|
|
chr7_+_18548963
|
1.147
|
|
HDAC9
|
histone deacetylase 9
|
|
chr10_+_22605300
|
1.145
|
NM_001204062
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough
COMM domain containing 3
|
|
chr11_-_85430117
|
1.143
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chrX_+_70765479
|
1.136
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr16_-_75272524
|
1.134
|
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr4_+_123161119
|
1.134
|
|
KIAA1109
|
KIAA1109
|
|
chr7_+_28530747
|
1.131
|
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr15_-_86338017
|
1.130
|
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr22_-_40859423
|
1.129
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr4_-_2263808
|
1.128
|
|
MXD4
|
MAX dimerization protein 4
|
|
chrM_+_2211
|
1.121
|
|
|
|
|
chr13_+_36050885
|
1.120
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr17_-_39092714
|
1.120
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr2_-_211018294
|
1.112
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr22_+_40440664
|
1.111
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr5_+_138678130
|
1.106
|
NM_001033112
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr7_+_17382900
|
1.103
|
|
AHR
|
aryl hydrocarbon receptor
|
|
chr9_+_96846710
|
1.099
|
NM_001253829
NM_152422
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr14_-_21572789
|
1.097
|
NM_001102454
|
ZNF219
|
zinc finger protein 219
|
|
chr12_-_86230158
|
1.094
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr1_+_201476283
|
1.090
|
|
|
|
|
chr11_-_46615584
|
1.089
|
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr19_+_48964505
|
1.088
|
NM_170720
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr2_+_171571936
|
1.087
|
|
|
|
|
chr1_+_167298382
|
1.077
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr2_-_208994408
|
1.077
|
NM_020989
|
CRYGC
|
crystallin, gamma C
|
|
chr17_-_46703613
|
1.076
|
NM_024017
|
HOXB9
|
homeobox B9
|