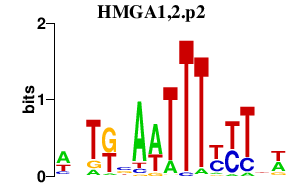
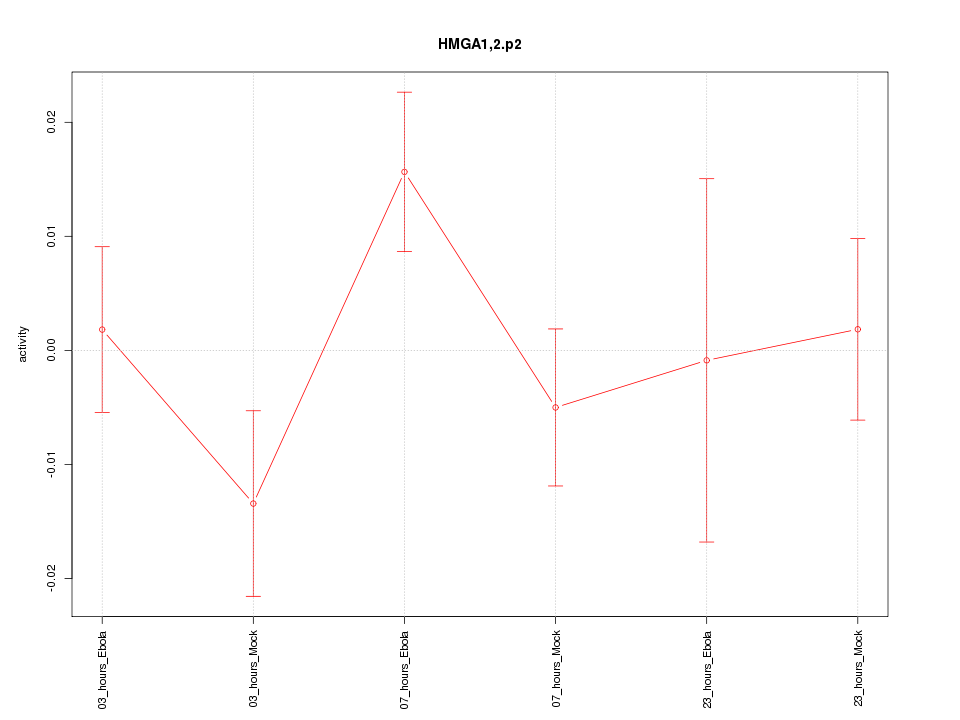
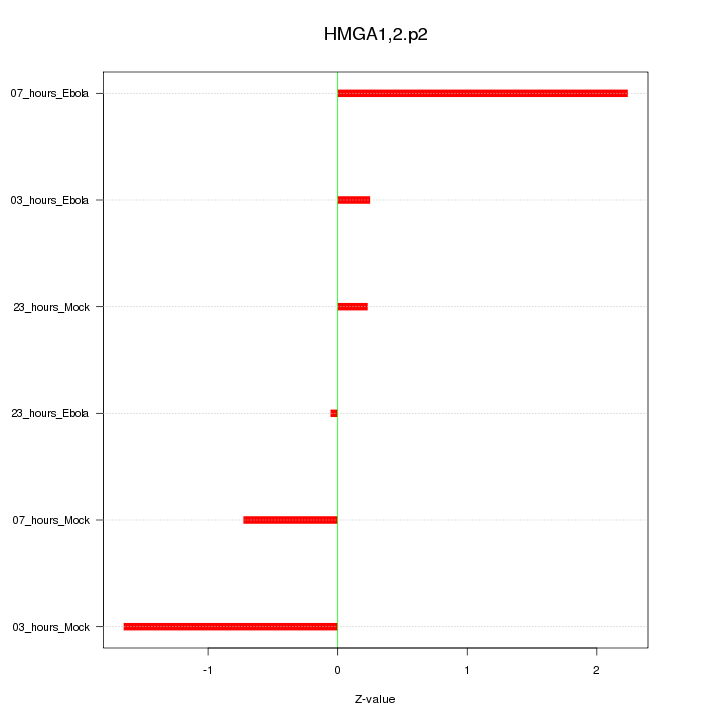
|
chr6_-_87804773
|
1.031
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr10_+_5238756
|
0.855
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr4_+_74347461
|
0.690
|
NM_001133
|
AFM
|
afamin
|
|
chr6_+_132873829
|
0.663
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr16_-_90096308
|
0.569
|
NM_001214
|
C16orf3
|
chromosome 16 open reading frame 3
|
|
chr3_-_167191808
|
0.543
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr6_+_149721494
|
0.479
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr11_+_56756346
|
0.478
|
NM_001005323
|
OR5AK2
|
olfactory receptor, family 5, subfamily AK, member 2
|
|
chr1_+_196857143
|
0.475
|
NM_001201550
NM_001201551
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr6_+_27925018
|
0.466
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr5_+_40909617
|
0.423
|
|
C7
|
complement component 7
|
|
chr3_-_42843217
|
0.423
|
|
|
|
|
chr5_-_172083569
|
0.421
|
|
|
|
|
chr11_-_85469043
|
0.411
|
NM_001162951
NM_001162953
|
SYTL2
|
synaptotagmin-like 2
|
|
chr6_+_150690027
|
0.401
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr11_-_18062304
|
0.393
|
NM_004179
|
TPH1
|
tryptophan hydroxylase 1
|
|
chr6_+_26020677
|
0.378
|
NM_003529
|
HIST1H3A
|
histone cluster 1, H3a
|
|
chr7_+_141490016
|
0.376
|
NM_018980
|
TAS2R5
|
taste receptor, type 2, member 5
|
|
chrX_-_23855456
|
0.372
|
|
RPL9
|
ribosomal protein L9
|
|
chr4_+_187187342
|
0.366
|
|
F11
|
coagulation factor XI
|
|
chr5_-_64777703
|
0.365
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr6_+_127898318
|
0.354
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr11_-_6817013
|
0.353
|
NM_003696
|
OR6A2
|
olfactory receptor, family 6, subfamily A, member 2
|
|
chr2_-_99917638
|
0.351
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr5_+_40909558
|
0.348
|
NM_000587
|
C7
|
complement component 7
|
|
chr4_+_156824846
|
0.343
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chrX_+_65384078
|
0.340
|
NM_001130860
NM_014799
|
HEPH
|
hephaestin
|
|
chr2_-_188312872
|
0.336
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr19_-_44384240
|
0.335
|
NM_001033719
|
ZNF404
|
zinc finger protein 404
|
|
chr5_+_175511848
|
0.320
|
NM_001079529
|
FAM153B
|
family with sequence similarity 153, member B
|
|
chr4_+_110749149
|
0.316
|
NM_006583
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chr6_-_68598987
|
0.316
|
|
ERVH-3
|
endogenous retrovirus group H, member 3
|
|
chr7_-_7758237
|
0.312
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr1_+_156030965
|
0.309
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr2_-_178787522
|
0.302
|
NM_001077358
|
PDE11A
|
phosphodiesterase 11A
|
|
chr1_-_161337663
|
0.293
|
NM_001013625
|
C1orf192
|
chromosome 1 open reading frame 192
|
|
chr11_-_5323174
|
0.286
|
NM_033179
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4
|
|
chr19_+_48964505
|
0.285
|
NM_170720
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr4_-_88450243
|
0.281
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr12_-_8218930
|
0.281
|
NM_004054
|
C3AR1
|
complement component 3a receptor 1
|
|
chr8_+_18248754
|
0.280
|
NM_000015
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase)
|
|
chr14_-_73453651
|
0.274
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr12_-_10022457
|
0.273
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chrX_-_21776158
|
0.260
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr4_+_88720701
|
0.259
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr1_+_79086087
|
0.257
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr2_+_177028804
|
0.257
|
NM_006898
|
HOXD3
|
homeobox D3
|
|
chr13_+_36050885
|
0.255
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr19_-_16250695
|
0.255
|
|
|
|
|
chr19_+_49109999
|
0.253
|
NM_133498
|
SPACA4
|
sperm acrosome associated 4
|
|
chr17_-_38859976
|
0.249
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr3_-_15563160
|
0.246
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr12_-_11244828
|
0.246
|
NM_176884
|
TAS2R43
TAS2R31
|
taste receptor, type 2, member 43
taste receptor, type 2, member 31
|
|
chr13_-_103346847
|
0.243
|
NM_001010977
|
METTL21C
|
methyltransferase like 21C
|
|
chr7_-_41742666
|
0.240
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chrX_-_62570968
|
0.238
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chrX_-_10645726
|
0.238
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_+_36065052
|
0.237
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr16_+_53479685
|
0.237
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr11_+_89392464
|
0.236
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr20_+_12989616
|
0.235
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr6_+_50786438
|
0.234
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr1_-_203317366
|
0.231
|
|
FMOD
|
fibromodulin
|
|
chr4_+_74270832
|
0.231
|
|
ALB
|
albumin
|
|
chr5_+_40909487
|
0.228
|
|
C7
|
complement component 7
|
|
chr1_-_148176392
|
0.226
|
|
|
|
|
chr11_+_124134699
|
0.221
|
NM_001005198
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5
|
|
chr11_+_27015668
|
0.218
|
|
|
|
|
chr3_+_171561138
|
0.218
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr1_-_244006471
|
0.217
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr7_+_141408152
|
0.215
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr17_+_48821106
|
0.215
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr12_+_100041527
|
0.208
|
NM_153364
|
FAM71C
|
family with sequence similarity 71, member C
|
|
chr6_-_116447225
|
0.207
|
NM_000493
|
COL10A1
|
collagen, type X, alpha 1
|
|
chr2_+_48807725
|
0.201
|
NM_001198594
|
STON1
STON1-GTF2A1L
|
stonin 1
STON1-GTF2A1L readthrough
|
|
chr9_+_92219926
|
0.199
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr2_-_175462428
|
0.198
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chrX_-_10535476
|
0.197
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr7_-_99474655
|
0.195
|
NM_001005276
|
OR2AE1
|
olfactory receptor, family 2, subfamily AE, member 1
|
|
chr20_-_17539480
|
0.193
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr12_-_113772900
|
0.192
|
NM_024959
|
SLC24A6
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 6
|
|
chr4_-_72649734
|
0.192
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr19_-_46526555
|
0.191
|
NM_005091
|
PGLYRP1
|
peptidoglycan recognition protein 1
|
|
chr1_+_84768333
|
0.189
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr2_-_224467049
|
0.189
|
|
SCG2
|
secretogranin II
|
|
chr5_-_176836535
|
0.187
|
NM_000505
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr11_-_76998462
|
0.186
|
NM_182833
|
GDPD4
|
glycerophosphodiester phosphodiesterase domain containing 4
|
|
chr10_+_77542518
|
0.183
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr4_-_164534656
|
0.182
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr19_+_37342552
|
0.180
|
NM_001242475
NM_001242476
|
ZNF345
|
zinc finger protein 345
|
|
chr2_-_198540583
|
0.180
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr11_+_27015833
|
0.180
|
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chr22_-_37164073
|
0.179
|
NM_001177702
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr1_+_198608136
|
0.177
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr14_-_57960432
|
0.177
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr6_+_54173202
|
0.176
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr12_+_20963637
|
0.175
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr9_-_95166827
|
0.175
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr1_+_16330730
|
0.173
|
NM_178840
|
C1orf64
|
chromosome 1 open reading frame 64
|
|
chr7_+_102553343
|
0.173
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr2_-_71062904
|
0.172
|
NM_015717
|
CD207
|
CD207 molecule, langerin
|
|
chr1_-_151826042
|
0.172
|
NM_182578
|
THEM5
|
thioesterase superfamily member 5
|
|
chr14_+_58906398
|
0.172
|
NM_001244193
|
KIAA0586
|
KIAA0586
|
|
chr20_-_23066828
|
0.171
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr18_+_32290195
|
0.170
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_+_74269971
|
0.170
|
NM_000477
|
ALB
|
albumin
|
|
chr1_+_192127591
|
0.168
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr12_+_75728418
|
0.168
|
NM_152779
|
GLIPR1L1
|
GLI pathogenesis-related 1 like 1
|
|
chr3_-_101039403
|
0.168
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr11_+_17332454
|
0.167
|
|
|
|
|
chr21_+_35736322
|
0.166
|
NM_172201
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2
|
|
chrX_-_34150427
|
0.166
|
NM_203408
|
FAM47A
|
family with sequence similarity 47, member A
|
|
chr1_+_40713572
|
0.165
|
NM_001008740
|
TMCO2
|
transmembrane and coiled-coil domains 2
|
|
chr11_-_124180661
|
0.165
|
NM_001002917
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1
|
|
chr3_+_38307297
|
0.164
|
NM_004256
|
SLC22A13
|
solute carrier family 22 (organic anion transporter), member 13
|
|
chr9_+_126773851
|
0.164
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr16_+_20775311
|
0.162
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr5_+_134303595
|
0.161
|
NM_178019
|
CATSPER3
|
cation channel, sperm associated 3
|
|
chr4_+_170581212
|
0.161
|
NM_001243374
|
CLCN3
|
chloride channel 3
|
|
chr12_-_371994
|
0.161
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr9_+_130026755
|
0.160
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr12_+_21679190
|
0.160
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr12_-_11091805
|
0.160
|
NM_023922
|
TAS2R14
|
taste receptor, type 2, member 14
|
|
chr7_-_108142934
|
0.157
|
|
PNPLA8
|
patatin-like phospholipase domain containing 8
|
|
chr7_-_27183225
|
0.157
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr5_-_160279045
|
0.157
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr1_+_171060017
|
0.157
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr11_-_124546198
|
0.156
|
NM_001199922
|
SIAE
|
sialic acid acetylesterase
|
|
chr5_-_176836488
|
0.155
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr11_+_120973374
|
0.155
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr15_-_65398578
|
0.154
|
NM_001163692
|
UBAP1L
|
ubiquitin associated protein 1-like
|
|
chr3_-_100712248
|
0.153
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr3_-_10334600
|
0.153
|
NM_001134944
NM_001134945
NM_001134946
|
GHRL
|
ghrelin/obestatin prepropeptide
|
|
chr4_-_38666429
|
0.150
|
|
FLJ13197
|
uncharacterized FLJ13197
|
|
chr17_+_41003184
|
0.149
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr6_-_51952368
|
0.149
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr4_-_2230957
|
0.148
|
NM_181808
|
POLN
|
polymerase (DNA directed) nu
|
|
chr9_+_34458810
|
0.147
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr12_-_11150473
|
0.146
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr9_+_131185129
|
0.146
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr4_+_124317884
|
0.145
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr7_+_80267782
|
0.145
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr4_+_154178525
|
0.143
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr17_-_62097937
|
0.143
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr7_-_156468369
|
0.143
|
|
|
|
|
chr4_-_139163222
|
0.142
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr12_-_81991959
|
0.141
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr4_+_72204769
|
0.141
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_-_67311773
|
0.140
|
NM_018672
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr10_-_5446781
|
0.140
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr16_-_21314371
|
0.140
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr7_-_140302311
|
0.140
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr16_-_21289637
|
0.139
|
|
CRYM
|
crystallin, mu
|
|
chr1_+_84630575
|
0.139
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chrX_-_77225134
|
0.137
|
NM_001029891
|
PGAM4
|
phosphoglycerate mutase family member 4
|
|
chr10_-_29754478
|
0.137
|
|
SVIL
|
supervillin
|
|
chr22_+_21821458
|
0.137
|
NM_001207052
|
TMEM191C
|
transmembrane protein 191C
|
|
chr7_-_96654128
|
0.137
|
NM_005221
|
DLX5
|
distal-less homeobox 5
|
|
chr1_+_101003692
|
0.136
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr22_-_18923963
|
0.136
|
NM_001195226
NM_016335
|
PRODH
|
proline dehydrogenase (oxidase) 1
|
|
chr4_+_186990308
|
0.136
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr11_+_94800316
|
0.135
|
|
SRSF8
|
serine/arginine-rich splicing factor 8
|
|
chr3_-_15540378
|
0.135
|
NM_080538
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr6_+_26383323
|
0.135
|
NM_001197238
NM_001197239
NM_001197240
NM_006995
NM_181531
NM_001197237
|
BTN2A2
|
butyrophilin, subfamily 2, member A2
|
|
chr1_-_162346579
|
0.134
|
NM_182581
|
C1orf111
|
chromosome 1 open reading frame 111
|
|
chr9_-_107666046
|
0.134
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr4_-_52883785
|
0.134
|
NM_001024611
|
LRRC66
|
leucine rich repeat containing 66
|
|
chr4_+_187070326
|
0.134
|
NM_001006655
|
FAM149A
|
family with sequence similarity 149, member A
|
|
chr9_+_5890801
|
0.133
|
NM_005511
|
MLANA
|
melan-A
|
|
chr14_-_21271002
|
0.133
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chrX_+_49832214
|
0.131
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr16_-_4524895
|
0.131
|
NM_020677
|
NMRAL1
|
NmrA-like family domain containing 1
|
|
chr18_+_66465316
|
0.130
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr1_+_104097321
|
0.130
|
NM_020978
|
AMY2B
|
amylase, alpha 2B (pancreatic)
|
|
chr19_+_10959105
|
0.130
|
NM_001136482
|
C19orf38
|
chromosome 19 open reading frame 38
|
|
chr8_+_39442086
|
0.128
|
NM_001190956
NM_014237
|
ADAM18
|
ADAM metallopeptidase domain 18
|
|
chr12_+_120740123
|
0.128
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr3_-_112564789
|
0.128
|
NM_001008784
NM_001199215
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr11_+_64808395
|
0.128
|
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr19_-_12992249
|
0.127
|
NM_001375
|
DNASE2
|
deoxyribonuclease II, lysosomal
|
|
chr11_+_66395733
|
0.127
|
|
|
|
|
chr12_-_14996381
|
0.126
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chr12_-_96336373
|
0.125
|
NM_182496
|
CCDC38
|
coiled-coil domain containing 38
|
|
chr7_+_134916730
|
0.124
|
NM_182489
|
STRA8
|
stimulated by retinoic acid gene 8 homolog (mouse)
|
|
chr9_+_125795787
|
0.124
|
|
RABGAP1
GPR21
|
RAB GTPase activating protein 1
G protein-coupled receptor 21
|
|
chr3_-_165555252
|
0.124
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr19_+_47813074
|
0.123
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr3_-_197300193
|
0.123
|
NM_203315
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr1_-_209957872
|
0.123
|
NM_152485
|
C1orf74
|
chromosome 1 open reading frame 74
|
|
chr7_-_99527242
|
0.122
|
NM_181538
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa
|
|
chr3_-_155524037
|
0.122
|
NM_173657
|
C3orf33
|
chromosome 3 open reading frame 33
|
|
chr14_-_74025643
|
0.122
|
NM_001220484
NM_203309
|
HEATR4
|
HEAT repeat containing 4
|
|
chr8_+_75736771
|
0.121
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr3_+_118892363
|
0.121
|
NM_006952
|
UPK1B
|
uroplakin 1B
|
|
chr22_+_21319417
|
0.121
|
NM_001018060
NM_144704
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr7_-_26232148
|
0.121
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr13_-_86373328
|
0.120
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr3_-_57530067
|
0.120
|
NM_178504
NM_198564
|
DNAH12
|
dynein, axonemal, heavy chain 12
|
|
chr10_+_695887
|
0.119
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr19_+_10397642
|
0.118
|
NM_001039132
NM_001544
NM_022377
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|