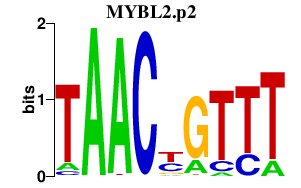
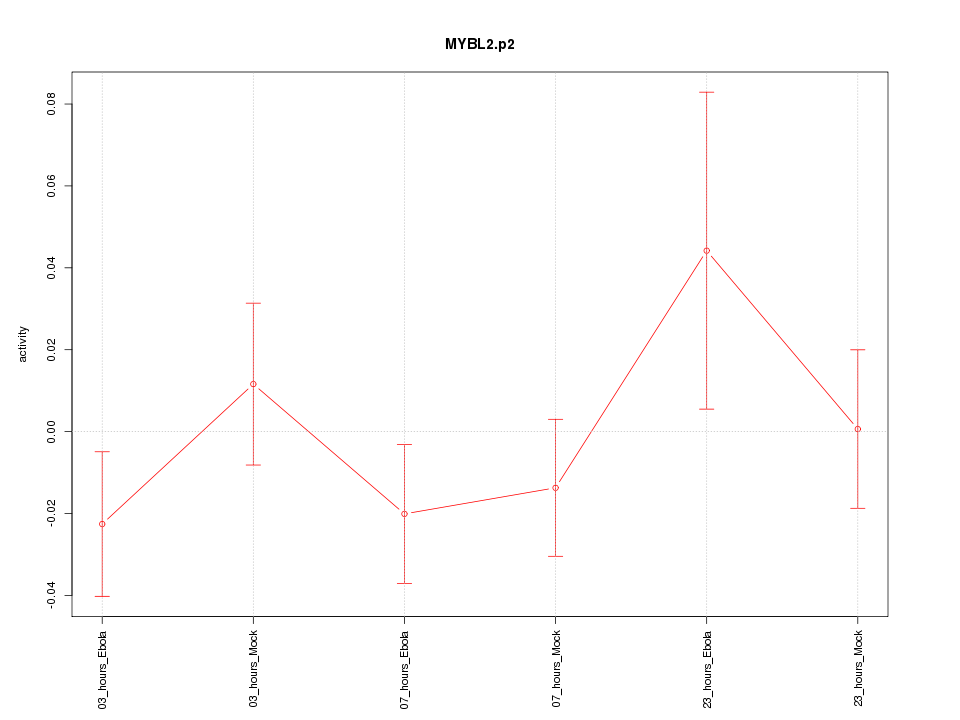
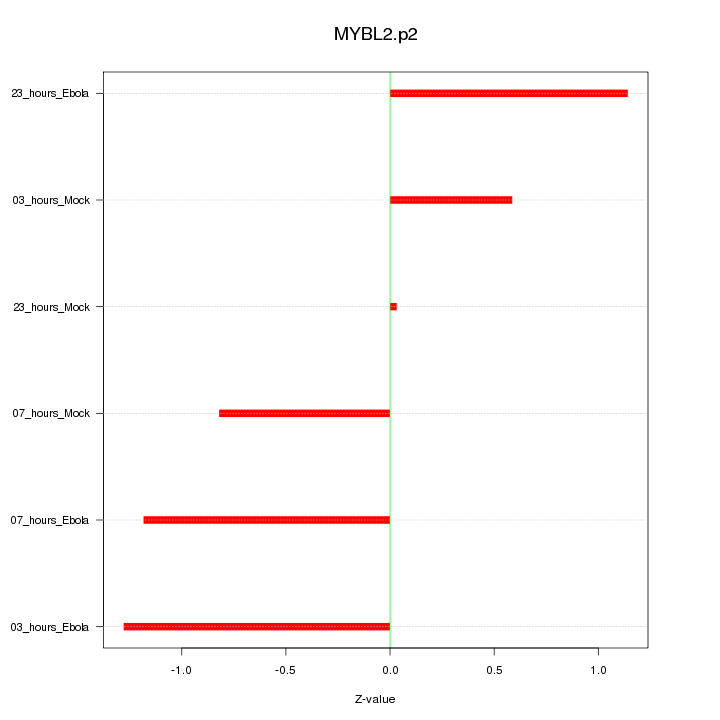
|
chr2_-_198540583
|
1.424
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr7_+_28530747
|
1.330
|
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_28452143
|
1.311
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_-_29738
|
1.271
|
NM_182905
|
WASH1
|
WAS protein family homolog 1
|
|
chr19_-_13947101
|
1.222
|
|
LOC284454
|
uncharacterized LOC284454
|
|
chr3_+_157154579
|
1.169
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr13_+_97999289
|
1.064
|
|
|
|
|
chr1_-_193155723
|
0.986
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr1_-_150780705
|
0.973
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr5_-_39424930
|
0.956
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr5_-_39425030
|
0.925
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_184243578
|
0.923
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr17_-_49124128
|
0.901
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr5_-_39425297
|
0.884
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr9_-_73029539
|
0.866
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr12_-_15865847
|
0.847
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr8_-_95274520
|
0.775
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr1_+_168545710
|
0.745
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr3_-_114478117
|
0.734
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr14_-_73453651
|
0.708
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr4_+_88571453
|
0.699
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr12_+_56915587
|
0.694
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr18_+_22040592
|
0.642
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr15_-_77154284
|
0.628
|
NM_001145923
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr2_+_29038862
|
0.619
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr2_-_152590983
|
0.614
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr8_-_95274540
|
0.613
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr3_+_141043054
|
0.610
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr17_+_7462064
|
0.570
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr22_-_39268169
|
0.560
|
|
CBX6
|
chromobox homolog 6
|
|
chr3_-_47933997
|
0.537
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr10_-_92680784
|
0.532
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr14_-_21567069
|
0.529
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chr12_-_26277807
|
0.525
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr7_+_18535351
|
0.521
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr22_-_39268214
|
0.512
|
|
CBX6
|
chromobox homolog 6
|
|
chr14_-_92198383
|
0.509
|
NM_024764
|
CATSPERB
|
cation channel, sperm-associated, beta
|
|
chr9_-_5968367
|
0.488
|
|
|
|
|
chrX_+_118108575
|
0.483
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr13_+_24144402
|
0.466
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr2_+_231281470
|
0.463
|
NM_001206704
|
SP100
|
SP100 nuclear antigen
|
|
chr7_+_104752312
|
0.455
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr4_-_83347778
|
0.431
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr19_+_48828820
|
0.430
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr15_+_57511616
|
0.429
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr9_+_96846710
|
0.426
|
NM_001253829
NM_152422
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr5_-_159846043
|
0.420
|
NM_006425
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae)
|
|
chr12_-_11002051
|
0.412
|
NM_001098538
NM_007244
|
PRR4
|
proline rich 4 (lacrimal)
|
|
chr22_+_40573879
|
0.398
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_+_141087298
|
0.396
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr16_+_69680959
|
0.392
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr4_+_71494460
|
0.391
|
NM_031889
|
ENAM
|
enamelin
|
|
chr10_-_32667543
|
0.386
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr21_+_30517897
|
0.381
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr15_-_25650647
|
0.378
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr1_+_148577356
|
0.377
|
NM_001102663
|
NBPF15
NBPF16
|
neuroblastoma breakpoint family, member 15
neuroblastoma breakpoint family, member 16
|
|
chr11_-_83984230
|
0.372
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chrX_+_118108595
|
0.372
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr16_+_24567628
|
0.364
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr2_+_210444154
|
0.361
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr12_-_57505121
|
0.358
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr3_+_63884074
|
0.354
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr7_-_100239136
|
0.348
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr10_-_92681025
|
0.347
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr10_+_17270257
|
0.345
|
NM_003380
|
VIM
|
vimentin
|
|
chr7_+_116395440
|
0.340
|
|
|
|
|
chr12_-_16757944
|
0.339
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_+_88720701
|
0.338
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr2_+_96905976
|
0.327
|
NM_001037228
|
LOC285033
|
uncharacterized LOC285033
|
|
chr2_-_208994408
|
0.326
|
NM_020989
|
CRYGC
|
crystallin, gamma C
|
|
chr10_+_17270469
|
0.323
|
|
VIM
|
vimentin
|
|
chr2_+_210444364
|
0.323
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr6_-_138189313
|
0.321
|
|
|
|
|
chr22_-_39268230
|
0.320
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr12_-_57504935
|
0.316
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr19_+_11200037
|
0.316
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr19_+_48828627
|
0.313
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr15_-_77176191
|
0.311
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr5_+_176873795
|
0.310
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr22_+_38071612
|
0.303
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr14_-_89016806
|
0.302
|
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr2_+_231280870
|
0.300
|
NM_001080391
NM_001206701
NM_001206702
NM_001206703
NM_003113
|
SP100
|
SP100 nuclear antigen
|
|
chr6_+_35265654
|
0.298
|
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr18_+_21032786
|
0.297
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr12_-_15114495
|
0.292
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr9_-_73028278
|
0.292
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr3_-_187452669
|
0.289
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr6_+_100054649
|
0.286
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr17_+_56833231
|
0.282
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr13_-_60325073
|
0.280
|
|
|
|
|
chr2_-_233641225
|
0.279
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr4_-_39979549
|
0.277
|
NM_001100399
NM_001100400
|
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae)
|
|
chr19_-_17414083
|
0.277
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr19_-_40948504
|
0.274
|
NM_013368
|
SERTAD3
|
SERTA domain containing 3
|
|
chr17_-_42908178
|
0.268
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr12_-_57505176
|
0.268
|
NM_001178078
NM_001178080
NM_001178081
NM_003153
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr3_+_121774208
|
0.265
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr3_-_114790221
|
0.262
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_+_32687246
|
0.262
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr7_-_93189121
|
0.258
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chr14_+_67656109
|
0.256
|
NM_173526
|
FAM71D
|
family with sequence similarity 71, member D
|
|
chr10_-_94003002
|
0.256
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr1_+_218518675
|
0.256
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr11_-_84634219
|
0.255
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_+_187930720
|
0.253
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_-_172858959
|
0.252
|
NM_031955
|
SPATA16
|
spermatogenesis associated 16
|
|
chr3_+_119316694
|
0.251
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr9_+_96846804
|
0.251
|
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr3_-_57530067
|
0.250
|
NM_178504
NM_198564
|
DNAH12
|
dynein, axonemal, heavy chain 12
|
|
chr12_-_57642922
|
0.249
|
|
|
|
|
chr16_-_52581713
|
0.246
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr13_-_20346398
|
0.246
|
|
PSPC1
|
paraspeckle component 1
|
|
chr18_+_8718448
|
0.245
|
|
|
|
|
chr2_+_37571752
|
0.238
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr11_+_62475165
|
0.237
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr13_-_101185944
|
0.237
|
NM_033110
|
A2LD1
|
AIG2-like domain 1
|
|
chr11_+_92085261
|
0.237
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr3_+_122334461
|
0.235
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr12_+_28410132
|
0.235
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr7_+_138144847
|
0.234
|
NM_003852
NM_015905
|
TRIM24
|
tripartite motif containing 24
|
|
chr6_-_90024936
|
0.233
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr12_-_15815073
|
0.227
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr2_+_228356287
|
0.226
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr3_-_44489718
|
0.223
|
|
|
|
|
chr11_+_100862719
|
0.223
|
NM_032021
|
TMEM133
|
transmembrane protein 133
|
|
chr3_+_187943192
|
0.221
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr11_-_85425790
|
0.220
|
NM_032379
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_+_6714717
|
0.219
|
NM_001171805
NM_006999
|
PAPD7
|
PAP associated domain containing 7
|
|
chr16_+_22103861
|
0.219
|
NM_173615
|
VWA3A
|
von Willebrand factor A domain containing 3A
|
|
chr6_+_135516879
|
0.219
|
|
|
|
|
chr11_+_65268313
|
0.214
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr14_+_65453947
|
0.213
|
|
|
|
|
chr11_-_6495688
|
0.211
|
NM_001248006
|
TRIM3
|
tripartite motif containing 3
|
|
chr3_-_25683825
|
0.209
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr10_+_5238756
|
0.207
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chrM_+_1670
|
0.207
|
|
|
|
|
chr20_+_47835964
|
0.204
|
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27
|
|
chr2_-_88926863
|
0.204
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr15_-_72563627
|
0.203
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr14_-_37641864
|
0.203
|
NM_001171170
NM_030631
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21
|
|
chr3_-_50608421
|
0.203
|
NM_001171741
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr20_+_47835954
|
0.202
|
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27
|
|
chr15_-_42749710
|
0.201
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr12_-_71031174
|
0.201
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr12_-_57351381
|
0.198
|
NM_003708
|
RDH16
|
retinol dehydrogenase 16 (all-trans)
|
|
chr17_+_18647325
|
0.198
|
NM_031456
|
FBXW10
|
F-box and WD repeat domain containing 10
|
|
chr16_+_81528953
|
0.196
|
NM_030629
|
CMIP
|
c-Maf inducing protein
|
|
chr19_-_16242996
|
0.195
|
|
|
|
|
chr22_-_41593466
|
0.194
|
|
|
|
|
chrX_-_19988381
|
0.193
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr2_+_231280906
|
0.193
|
|
SP100
|
SP100 nuclear antigen
|
|
chr6_+_87967433
|
0.191
|
|
ZNF292
|
zinc finger protein 292
|
|
chr21_+_38792600
|
0.190
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr1_+_22778347
|
0.189
|
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr11_-_85430117
|
0.189
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr2_+_231280933
|
0.188
|
|
SP100
|
SP100 nuclear antigen
|
|
chr6_-_43655539
|
0.184
|
NM_001193343
NM_018135
|
MRPS18A
|
mitochondrial ribosomal protein S18A
|
|
chr7_+_112120907
|
0.183
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr14_-_78174320
|
0.182
|
NM_006020
|
ALKBH1
|
alkB, alkylation repair homolog 1 (E. coli)
|
|
chr5_-_41213513
|
0.180
|
NM_000065
|
C6
|
complement component 6
|
|
chr11_+_62475113
|
0.179
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr8_+_32579350
|
0.179
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr3_+_16974581
|
0.178
|
NM_015184
|
PLCL2
|
phospholipase C-like 2
|
|
chr4_-_47840058
|
0.178
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr2_-_134326030
|
0.177
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr17_-_48227873
|
0.177
|
NM_032595
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B
|
|
chr6_-_117747017
|
0.176
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr1_-_154832187
|
0.174
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr12_-_14967091
|
0.173
|
NM_001013698
|
C12orf69
|
chromosome 12 open reading frame 69
|
|
chr1_-_46089638
|
0.172
|
NM_001114938
NM_001190182
|
CCDC17
|
coiled-coil domain containing 17
|
|
chr11_+_33563772
|
0.171
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr11_+_61102035
|
0.171
|
|
DAK
|
dihydroxyacetone kinase 2 homolog (S. cerevisiae)
|
|
chr6_-_108145507
|
0.169
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr19_+_19144617
|
0.169
|
|
ARMC6
|
armadillo repeat containing 6
|
|
chr14_-_69260630
|
0.169
|
NM_001244698
NM_004926
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr3_-_167371288
|
0.169
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr12_+_80838125
|
0.167
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr4_+_39063851
|
0.165
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr3_-_50378238
|
0.164
|
NM_001206957
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr16_+_31470104
|
0.163
|
NM_001105247
NM_024742
|
ARMC5
|
armadillo repeat containing 5
|
|
chrX_+_105412297
|
0.162
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr3_+_180630054
|
0.160
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr18_+_32335939
|
0.159
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_-_39979159
|
0.159
|
|
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae)
|
|
chr7_+_28725597
|
0.159
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_-_67579478
|
0.159
|
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr12_-_121248679
|
0.159
|
|
SPPL3
|
signal peptide peptidase-like 3
|
|
chr10_-_74283693
|
0.158
|
NM_001195519
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr1_-_149908098
|
0.158
|
NM_181873
|
MTMR11
|
myotubularin related protein 11
|
|
chr16_+_30773619
|
0.157
|
|
RNF40
|
ring finger protein 40
|
|
chr15_-_60919642
|
0.157
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chrX_-_128788913
|
0.156
|
NM_017413
|
APLN
|
apelin
|
|
chr10_+_114154522
|
0.155
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr2_+_138721807
|
0.154
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr14_-_21566728
|
0.154
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr1_-_155649597
|
0.154
|
NM_001198899
|
YY1AP1
|
YY1 associated protein 1
|
|
chr15_-_58306185
|
0.153
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr2_-_216245573
|
0.152
|
|
FN1
|
fibronectin 1
|
|
chr9_-_100700422
|
0.151
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr10_+_47894022
|
0.151
|
NM_018232
|
FAM21B
|
family with sequence similarity 21, member B
|