|
chr19_-_1237840
|
0.920
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr1_+_212788359
|
0.720
|
NM_001206486
|
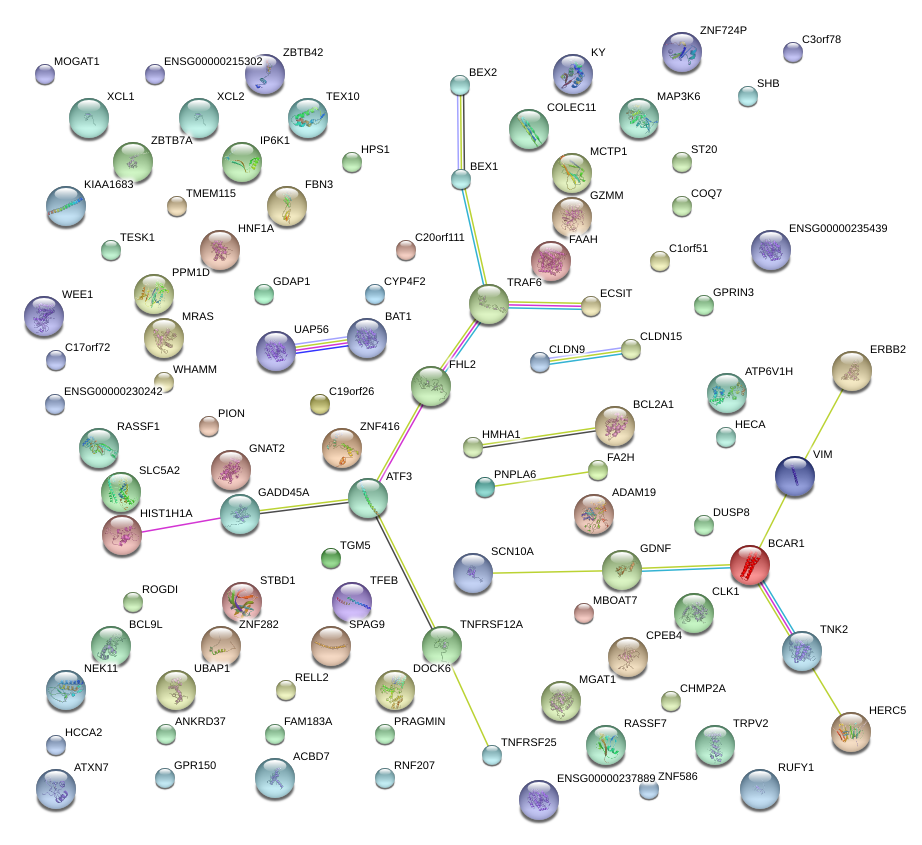
ATF3
|
activating transcription factor 3
|
|
chr11_+_61735217
|
0.633
|
|
LOC399900
|
uncharacterized LOC399900
|
|
chr5_-_37839781
|
0.579
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr1_+_43613593
|
0.514
|
NM_001101376
|
FAM183A
|
family with sequence similarity 183, member A
|
|
chr16_+_3062456
|
0.454
|
NM_020982
|
CLDN9
|
claudin 9
|
|
chr15_-_80215663
|
0.449
|
NM_001100880
NM_001199757
|
ST20
|
suppressor of tumorigenicity 20
|
|
chr4_+_186317964
|
0.440
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr10_+_17271956
|
0.371
|
|
VIM
|
vimentin
|
|
chr1_+_212738675
|
0.353
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr1_-_6526150
|
0.346
|
NM_001039664
NM_003790
NM_148965
NM_148966
NM_148967
NM_148970
|
TNFRSF25
|
tumor necrosis factor receptor superfamily, member 25
|
|
chr4_+_186317837
|
0.342
|
NM_181726
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr15_+_83478329
|
0.337
|
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chrX_+_124453968
|
0.328
|
NM_001195272
|
LOC100129520
|
testis expressed sequence 13-like
|
|
chr4_+_89378234
|
0.316
|
NM_016323
|
HERC5
|
hect domain and RLD 5
|
|
chr16_+_3070310
|
0.310
|
NM_016639
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A
|
|
chr5_+_141016567
|
0.309
|
|
RELL2
|
RELT-like 2
|
|
chr19_-_16008870
|
0.309
|
NM_001082
|
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2
|
|
chr1_+_6266140
|
0.302
|
NM_207396
|
RNF207
|
ring finger protein 207
|
|
chr4_+_186317892
|
0.298
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr5_+_141016987
|
0.295
|
|
RELL2
|
RELT-like 2
|
|
chr8_-_8239188
|
0.294
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr5_+_173315313
|
0.292
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr6_-_41703345
|
0.282
|
|
TFEB
|
transcription factor EB
|
|
chr19_-_4065467
|
0.272
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr10_-_17271955
|
0.266
|
|
|
|
|
chr17_+_62075710
|
0.264
|
NM_001164257
NM_001191029
NM_001191030
NM_001191031
|
C17orf72
|
chromosome 17 open reading frame 72
|
|
chr10_-_100206643
|
0.263
|
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr22_-_41593466
|
0.260
|
|
|
|
|
chr19_-_11373117
|
0.254
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr5_+_178986692
|
0.249
|
NM_001040451
NM_001040452
|
RUFY1
|
RUN and FYVE domain containing 1
|
|
chr19_-_8212367
|
0.236
|
NM_032447
|
FBN3
|
fibrillin 3
|
|
chr2_-_106015423
|
0.232
|
NM_001039492
NM_001450
NM_201555
|
FHL2
|
four and a half LIM domains 2
|
|
chrX_-_119379060
|
0.230
|
|
NKAPP1
|
NFKB activating protein pseudogene 1
|
|
chr17_+_16318911
|
0.229
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr9_+_34179002
|
0.224
|
NM_001171201
NM_001171202
NM_001171203
NM_001171204
NM_016525
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr10_+_65224937
|
0.224
|
|
LOC84989
|
uncharacterized LOC84989
|
|
chr6_-_26017960
|
0.223
|
NM_005325
|
HIST1H1A
|
histone cluster 1, H1a
|
|
chr19_+_544026
|
0.221
|
NM_005317
|
GZMM
|
granzyme M (lymphocyte met-ase 1)
|
|
chr9_+_34179023
|
0.221
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr8_-_54755836
|
0.220
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr9_+_34179036
|
0.220
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr9_+_34179058
|
0.219
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr6_-_41702734
|
0.219
|
NM_007162
|
TFEB
|
transcription factor EB
|
|
chr10_+_17271859
|
0.215
|
|
VIM
|
vimentin
|
|
chr19_-_18385166
|
0.215
|
NM_001145304
NM_001145305
NM_025249
|
KIAA1683
|
KIAA1683
|
|
chr5_-_157002739
|
0.215
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr1_+_68150858
|
0.212
|
NM_001199741
NM_001199742
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr11_+_560868
|
0.212
|
NM_001143993
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr8_-_54755546
|
0.212
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr15_+_83477972
|
0.211
|
NM_001080435
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chr17_+_37844325
|
0.210
|
NM_001005862
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr8_+_75262616
|
0.209
|
NM_018972
NM_001040875
|
GDAP1
|
ganglioside-induced differentiation-associated protein 1
|
|
chr1_-_110155672
|
0.208
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr8_-_54755821
|
0.207
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr4_+_77227799
|
0.205
|
|
STBD1
|
starch binding domain 1
|
|
chr5_+_94955979
|
0.201
|
NM_199243
|
GPR150
|
G protein-coupled receptor 150
|
|
chr16_+_31494438
|
0.200
|
NM_003041
|
SLC5A2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2
|
|
chr16_-_75272979
|
0.199
|
NM_001170721
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr6_-_31509769
|
0.198
|
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B
|
|
chr2_-_201729241
|
0.198
|
NM_001162407
|
CLK1
|
CDC-like kinase 1
|
|
chr7_+_148892553
|
0.196
|
NM_003575
|
ZNF282
|
zinc finger protein 282
|
|
chr3_-_50374795
|
0.196
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr19_-_59066372
|
0.196
|
|
CHMP2A
|
charged multivesicular body protein 2A
|
|
chr1_+_1260289
|
0.194
|
|
|
|
|
chr15_-_43559054
|
0.194
|
NM_004245
NM_201631
|
TGM5
|
transglutaminase 5
|
|
chr19_-_11639679
|
0.191
|
NM_001243204
|
ECSIT
|
ECSIT homolog (Drosophila)
|
|
chr11_-_118781612
|
0.190
|
NM_182557
|
BCL9L
|
B-cell CLL/lymphoma 9-like
|
|
chr19_-_54693167
|
0.188
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr9_+_34179047
|
0.188
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr3_+_63953419
|
0.187
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr1_+_150255228
|
0.186
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr11_-_1507810
|
0.184
|
NM_001172223
|
MOB2
|
MOB kinase activator 2
|
|
chr20_-_42839295
|
0.184
|
|
C20orf111
|
chromosome 20 open reading frame 111
|
|
chr1_+_168545710
|
0.183
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr1_-_27692825
|
0.182
|
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr7_-_100881030
|
0.182
|
NM_014343
|
CLDN15
|
claudin 15
|
|
chr7_-_77045706
|
0.182
|
NM_017439
|
PION
|
pigeon homolog (Drosophila)
|
|
chr3_+_52570620
|
0.181
|
NM_001124767
|
C3orf78
|
chromosome 3 open reading frame 78
|
|
chr5_-_180230047
|
0.178
|
NM_002406
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr19_-_58090254
|
0.177
|
|
ZNF416
|
zinc finger protein 416
|
|
chr11_-_36531767
|
0.176
|
NM_004620
NM_145803
|
TRAF6
|
TNF receptor-associated factor 6
|
|
chr16_-_4852571
|
0.175
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr3_+_138067444
|
0.174
|
NM_001085049
NM_001252093
|
MRAS
|
muscle RAS oncogene homolog
|
|
chrX_-_102319096
|
0.174
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chr6_-_31509725
|
0.172
|
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B
|
|
chr10_-_15130773
|
0.170
|
NM_001039844
|
ACBD7
|
acyl-CoA binding domain containing 7
|
|
chr19_+_1067293
|
0.169
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr3_-_50374749
|
0.167
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr3_-_49823953
|
0.165
|
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr9_+_35605339
|
0.164
|
|
TESK1
|
testis-specific kinase 1
|
|
chr12_+_121416548
|
0.162
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr3_-_195619451
|
0.162
|
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr11_+_9596233
|
0.160
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr3_-_50396892
|
0.159
|
|
TMEM115
|
transmembrane protein 115
|
|
chr9_+_35605275
|
0.157
|
NM_006285
|
TESK1
|
testis-specific kinase 1
|
|
chr11_-_1593149
|
0.157
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr19_-_58090071
|
0.156
|
NM_017879
|
ZNF416
|
zinc finger protein 416
|
|
chr14_+_105266878
|
0.155
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr17_+_58677489
|
0.155
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr1_+_46859918
|
0.155
|
NM_001441
|
FAAH
|
fatty acid amide hydrolase
|
|
chr3_+_130745693
|
0.154
|
NM_001146003
NM_024800
NM_145910
|
NEK11
|
NIMA (never in mitosis gene a)- related kinase 11
|
|
chr1_+_16259046
|
0.153
|
|
|
|
|
chr19_+_7599037
|
0.153
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr17_-_49198084
|
0.152
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr9_-_38069082
|
0.151
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr4_-_90228943
|
0.150
|
|
GPRIN3
|
GPRIN family member 3
|
|
chr4_+_77227178
|
0.148
|
NM_003943
|
STBD1
|
starch binding domain 1
|
|
chr6_+_139456240
|
0.148
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr1_+_31885962
|
0.147
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr2_-_201729351
|
0.146
|
|
CLK1
|
CDC-like kinase 1
|
|
chr15_+_82555101
|
0.144
|
NM_001008226
|
FAM154B
|
family with sequence similarity 154, member B
|
|
chr19_-_45926819
|
0.143
|
NM_202001
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr12_+_3068442
|
0.142
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chrX_-_53024642
|
0.142
|
NM_001242489
NM_001242490
NM_001242492
NM_001242493
NM_001242494
NM_001242495
NM_001242496
NM_001242497
NM_014138
|
FAM156A
|
family with sequence similarity 156, member A
|
|
chr13_-_60738078
|
0.141
|
NM_001042517
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr7_+_143013218
|
0.141
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr1_+_43996478
|
0.139
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr19_+_48111437
|
0.139
|
NM_015711
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1
|
|
chr3_+_50304989
|
0.137
|
NM_001005914
NM_004636
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr5_+_76114881
|
0.137
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr11_+_71935824
|
0.135
|
NM_001567
|
INPPL1
|
inositol polyphosphate phosphatase-like 1
|
|
chr3_-_49823972
|
0.133
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr11_-_441995
|
0.131
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr21_-_33104387
|
0.131
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr19_+_1067536
|
0.130
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr19_+_54369610
|
0.130
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_-_44008984
|
0.130
|
NM_198850
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3
|
|
chr12_+_56324945
|
0.130
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr19_-_45926759
|
0.129
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr14_+_105267378
|
0.128
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr18_+_23806385
|
0.127
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chrX_+_153770458
|
0.127
|
NM_001099856
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr20_-_42839423
|
0.127
|
NM_016470
|
C20orf111
|
chromosome 20 open reading frame 111
|
|
chr19_-_3028901
|
0.126
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr19_-_11639929
|
0.126
|
NM_001142464
NM_001142465
NM_016581
|
ECSIT
|
ECSIT homolog (Drosophila)
|
|
chr11_-_441956
|
0.125
|
|
ANO9
|
anoctamin 9
|
|
chr15_-_72668184
|
0.125
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr3_-_194354009
|
0.125
|
NM_001011655
NM_001166305
NM_001166306
NM_138399
|
TMEM44
|
transmembrane protein 44
|
|
chr6_+_30851860
|
0.125
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr19_+_57791794
|
0.124
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr21_-_33104405
|
0.123
|
NM_001145444
NM_001145445
NM_020706
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr5_-_157002705
|
0.121
|
|
|
|
|
chr19_+_56124958
|
0.120
|
NM_001195605
|
ZNF865
|
zinc finger protein 865
|
|
chr13_+_97794050
|
0.119
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr9_+_132597651
|
0.119
|
NM_001110303
NM_006676
|
USP20
|
ubiquitin specific peptidase 20
|
|
chr22_+_42229107
|
0.119
|
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr2_-_201729377
|
0.118
|
NM_004071
|
CLK1
|
CDC-like kinase 1
|
|
chr5_+_141016516
|
0.117
|
NM_001130029
NM_173828
|
RELL2
|
RELT-like 2
|
|
chr8_-_109799769
|
0.116
|
NM_153015
|
TMEM74
|
transmembrane protein 74
|
|
chr2_+_223536362
|
0.116
|
NM_058165
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1
|
|
chr19_-_41255827
|
0.114
|
NM_198476
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr16_+_3014216
|
0.114
|
NM_001253725
NM_001253726
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr12_+_133195365
|
0.113
|
NM_012226
NM_016318
NM_170682
NM_170683
NM_174872
NM_174873
|
P2RX2
|
purinergic receptor P2X, ligand-gated ion channel, 2
|
|
chr5_-_68665316
|
0.113
|
NM_003187
NM_016283
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa
|
|
chr13_-_60737899
|
0.113
|
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr6_-_31550065
|
0.112
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr12_-_98897622
|
0.112
|
|
LOC643770
|
uncharacterized LOC643770
|
|
chr19_+_1067114
|
0.112
|
NM_012292
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr5_-_179229874
|
0.111
|
NM_054013
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr4_-_2935650
|
0.110
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr15_+_67358161
|
0.109
|
|
SMAD3
|
SMAD family member 3
|
|
chr3_+_138067678
|
0.109
|
NM_001252092
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr8_+_104383785
|
0.109
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr17_-_70588942
|
0.108
|
|
LOC100499467
|
uncharacterized LOC100499467
|
|
chr11_+_9685650
|
0.108
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr7_-_80548322
|
0.107
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr9_-_130742803
|
0.106
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr19_-_54693414
|
0.105
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr3_-_194353908
|
0.105
|
|
TMEM44
|
transmembrane protein 44
|
|
chr12_+_50017340
|
0.105
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr3_-_49823626
|
0.104
|
NM_001242829
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr11_+_17298278
|
0.104
|
NM_005013
|
NUCB2
|
nucleobindin 2
|
|
chr1_-_184943673
|
0.104
|
NM_052966
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr8_-_33424642
|
0.104
|
NM_024787
|
RNF122
|
ring finger protein 122
|
|
chr3_+_112709777
|
0.103
|
|
GTPBP8
|
GTP-binding protein 8 (putative)
|
|
chr6_-_31745048
|
0.103
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr4_-_185747051
|
0.103
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr19_-_45927081
|
0.102
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr4_+_77227882
|
0.102
|
|
STBD1
|
starch binding domain 1
|
|
chr17_+_58677603
|
0.102
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr17_+_1627833
|
0.100
|
NM_001163811
|
WDR81
|
WD repeat domain 81
|
|
chr1_+_109234908
|
0.100
|
NM_018061
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr2_-_85641128
|
0.100
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr8_+_145086444
|
0.100
|
NM_001134374
NM_198572
|
SPATC1
|
spermatogenesis and centriole associated 1
|
|
chr1_+_20915440
|
0.099
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr1_+_228328003
|
0.099
|
|
GUK1
|
guanylate kinase 1
|
|
chr6_-_28186663
|
0.099
|
|
TOB2P1
|
transducer of ERBB2, 2 pseudogene 1
|
|
chr1_+_231298673
|
0.098
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr6_+_35420115
|
0.098
|
NM_021922
|
FANCE
|
Fanconi anemia, complementation group E
|
|
chr7_-_32931189
|
0.098
|
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr1_-_2322990
|
0.098
|
NM_024848
|
MORN1
|
MORN repeat containing 1
|
|
chr17_-_4890664
|
0.097
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr5_-_43040147
|
0.097
|
NM_001014279
|
C5orf39
|
chromosome 5 open reading frame 39
|
|
chr13_+_60970590
|
0.097
|
NM_001146071
NM_030794
|
TDRD3
|
tudor domain containing 3
|
|
chr20_-_32262254
|
0.097
|
NM_031231
NM_031232
|
NECAB3
|
N-terminal EF-hand calcium binding protein 3
|
|
chr5_-_180229713
|
0.096
|
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr4_-_76555570
|
0.096
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr11_+_64851691
|
0.096
|
NM_006782
|
ZFPL1
|
zinc finger protein-like 1
|
|
chrX_-_153744372
|
0.096
|
NM_001171132
NM_001171133
NM_001171134
NM_021806
|
FAM3A
|
family with sequence similarity 3, member A
|