|
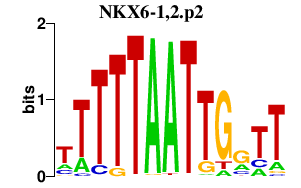
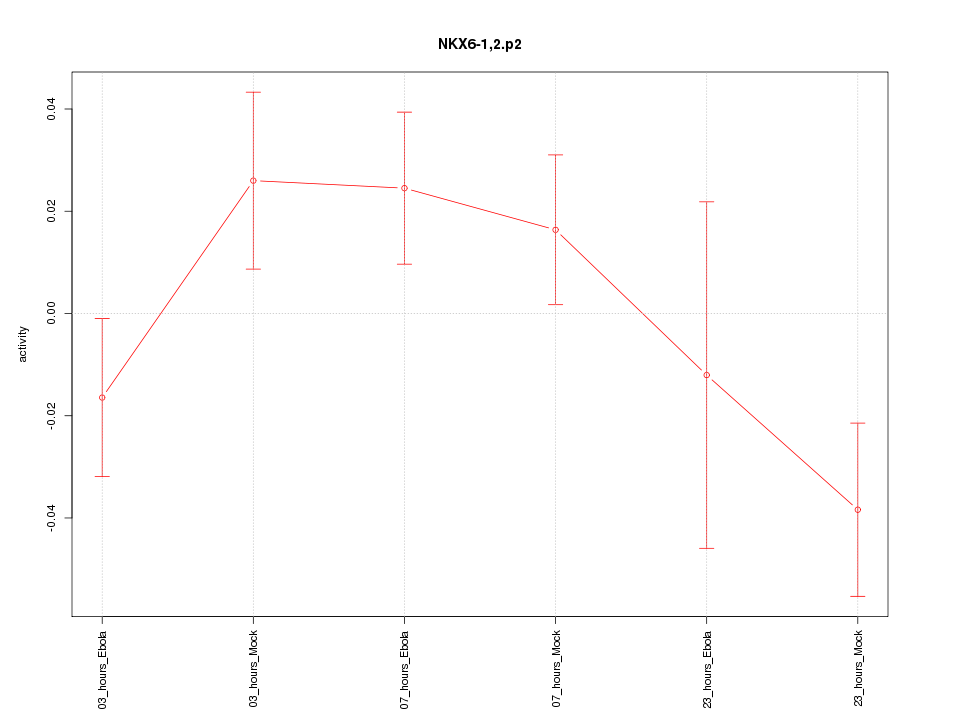
chr13_+_27844463
|
1.237
|
NM_206827
|
RASL11A
|
RAS-like, family 11, member A
|
|
chr6_-_26043884
|
1.125
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr8_-_82607206
|
0.925
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr11_-_71524890
|
0.783
|
|
LOC285407
|
asparagine-linked glycosylation 1 homolog pseudogene
|
|
chr5_+_38445577
|
0.736
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr1_-_150738292
|
0.732
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr6_+_3259134
|
0.701
|
NM_001128591
NM_001128592
NM_001135750
|
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4
|
|
chrX_-_23855456
|
0.700
|
|
RPL9
|
ribosomal protein L9
|
|
chr1_-_150738232
|
0.646
|
|
CTSS
|
cathepsin S
|
|
chr1_+_182419255
|
0.599
|
NM_001137669
|
RGSL1
|
regulator of G-protein signaling like 1
|
|
chr1_-_150738255
|
0.588
|
|
CTSS
|
cathepsin S
|
|
chr21_-_31852631
|
0.585
|
NM_181607
|
KRTAP19-1
|
keratin associated protein 19-1
|
|
chr5_+_81601165
|
0.581
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr7_+_94297448
|
0.558
|
|
PEG10
|
paternally expressed 10
|
|
chr4_+_88896899
|
0.541
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr13_+_27844936
|
0.539
|
|
RASL11A
|
RAS-like, family 11, member A
|
|
chr2_+_210444364
|
0.535
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr4_-_70725792
|
0.532
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr2_-_228244005
|
0.528
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr4_+_151503076
|
0.500
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr5_+_31193852
|
0.500
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr20_-_18477811
|
0.495
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr16_+_28565225
|
0.491
|
NM_138414
|
CCDC101
|
coiled-coil domain containing 101
|
|
chr1_-_54483802
|
0.485
|
NM_001010978
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr11_-_125550674
|
0.485
|
NM_001612
NM_020069
NM_020107
NM_020108
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chr2_+_210444154
|
0.479
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr10_-_52008312
|
0.478
|
NM_001143974
NM_019893
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2
|
|
chr3_-_138048727
|
0.463
|
NM_178130
|
NME9
|
NME gene family member 9
|
|
chr4_+_74347461
|
0.453
|
NM_001133
|
AFM
|
afamin
|
|
chr5_+_31193761
|
0.449
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr6_+_5030202
|
0.448
|
|
LOC100129461
|
uncharacterized LOC100129461
|
|
chr12_+_32654976
|
0.442
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr1_+_186798031
|
0.435
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr7_-_102283237
|
0.425
|
NM_001114403
|
UPK3BL
|
uroplakin 3B-like
|
|
chr21_-_36421195
|
0.406
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr10_-_96829074
|
0.403
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr8_+_79428335
|
0.398
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr13_+_24144402
|
0.390
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr5_-_111091947
|
0.366
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_170115187
|
0.362
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr12_+_25205488
|
0.356
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_+_54410893
|
0.349
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr4_+_88896801
|
0.349
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr7_-_92855780
|
0.326
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr5_+_140729723
|
0.325
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr7_-_107880492
|
0.321
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chrX_+_36254050
|
0.321
|
NM_001098843
|
CXorf30
|
chromosome X open reading frame 30
|
|
chr14_-_57960432
|
0.320
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr3_-_79817058
|
0.315
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr18_-_51690988
|
0.314
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr21_-_36421578
|
0.310
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_+_132859426
|
0.306
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr11_-_62521533
|
0.304
|
NM_024784
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
|
chr5_+_68513624
|
0.301
|
|
MRPS36
|
mitochondrial ribosomal protein S36
|
|
chr5_-_58295758
|
0.301
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_+_68071386
|
0.292
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr19_-_46526555
|
0.290
|
NM_005091
|
PGLYRP1
|
peptidoglycan recognition protein 1
|
|
chrX_-_33229428
|
0.286
|
NM_004006
|
DMD
|
dystrophin
|
|
chr17_+_68100994
|
0.282
|
NM_170742
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr6_+_39760138
|
0.279
|
|
|
|
|
chr4_-_70518964
|
0.274
|
NM_001252274
NM_001252275
NM_006798
|
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus
|
|
chr6_+_53883713
|
0.273
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr6_+_26199712
|
0.271
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr5_+_149997205
|
0.271
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr21_-_36421630
|
0.271
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr21_-_31874407
|
0.266
|
NM_181611
|
KRTAP19-5
|
keratin associated protein 19-5
|
|
chr8_+_132991171
|
0.265
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr1_+_168545710
|
0.261
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr12_-_10573041
|
0.261
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr13_+_103459495
|
0.260
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr18_-_46477080
|
0.258
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr21_-_36421586
|
0.251
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_-_24690740
|
0.249
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr12_+_21284127
|
0.248
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr5_+_140566655
|
0.248
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr2_-_178787522
|
0.248
|
NM_001077358
|
PDE11A
|
phosphodiesterase 11A
|
|
chr6_-_146056896
|
0.247
|
NM_001018041
NM_005670
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin)
|
|
chr1_+_147400498
|
0.246
|
NM_016334
|
GPR89B
GPR89A
|
G protein-coupled receptor 89B
G protein-coupled receptor 89A
|
|
chr1_+_61869699
|
0.234
|
|
NFIA
|
nuclear factor I/A
|
|
chr8_+_9953061
|
0.232
|
NM_001135671
NM_001199729
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr21_-_36421461
|
0.231
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_-_170077807
|
0.231
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr10_-_115423640
|
0.231
|
NM_006175
NM_198060
|
NRAP
|
nebulin-related anchoring protein
|
|
chr6_+_114178516
|
0.231
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr19_-_49220213
|
0.230
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr6_-_133084585
|
0.229
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr6_-_135271206
|
0.225
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr7_-_41742666
|
0.225
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr3_-_71294229
|
0.225
|
NM_001244814
|
FOXP1
|
forkhead box P1
|
|
chr6_-_49712055
|
0.224
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr2_+_211342405
|
0.222
|
NM_001122633
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr1_-_17766142
|
0.217
|
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr5_+_140782367
|
0.217
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr20_+_23345020
|
0.217
|
NM_022482
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr17_-_38821286
|
0.214
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr14_+_74083547
|
0.214
|
NM_001037162
|
ACOT6
|
acyl-CoA thioesterase 6
|
|
chr10_+_72163899
|
0.213
|
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr13_-_47470967
|
0.211
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr3_-_151987332
|
0.210
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr4_+_88896868
|
0.208
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr6_+_132873829
|
0.208
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr4_+_88896838
|
0.207
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr18_-_24722757
|
0.204
|
NM_001243848
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr17_-_56494889
|
0.203
|
|
RNF43
|
ring finger protein 43
|
|
chr6_+_26402464
|
0.202
|
NM_001145008
NM_001145009
NM_007048
NM_194441
|
BTN3A1
|
butyrophilin, subfamily 3, member A1
|
|
chr3_-_168865521
|
0.198
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr4_+_15704752
|
0.191
|
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr19_-_48759141
|
0.190
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr14_-_60097314
|
0.190
|
|
RTN1
|
reticulon 1
|
|
chr10_+_74870209
|
0.189
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr1_+_84647342
|
0.189
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_-_231084600
|
0.188
|
NM_004509
NM_004510
NM_080424
|
SP110
|
SP110 nuclear body protein
|
|
chr1_+_145477018
|
0.187
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr14_-_95599794
|
0.186
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_+_102508333
|
0.185
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr10_-_113933517
|
0.185
|
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr9_-_77643307
|
0.184
|
NM_152420
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr5_+_42565963
|
0.183
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr14_-_71001731
|
0.182
|
NM_003814
|
ADAM20
|
ADAM metallopeptidase domain 20
|
|
chr3_+_49547867
|
0.182
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr12_-_54653369
|
0.180
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr18_+_72922725
|
0.179
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr12_-_109458844
|
0.179
|
NM_018711
|
SVOP
|
SV2 related protein homolog (rat)
|
|
chr12_-_54653317
|
0.178
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr2_-_74382352
|
0.178
|
|
|
|
|
chr19_-_41812991
|
0.176
|
|
|
|
|
chr1_+_12403045
|
0.176
|
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae)
|
|
chr2_-_183387063
|
0.176
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr6_-_138820580
|
0.174
|
|
|
|
|
chr1_+_192605267
|
0.173
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr2_+_202047803
|
0.172
|
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr11_-_124190092
|
0.171
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr2_+_183608329
|
0.168
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr7_+_28725597
|
0.167
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_140800536
|
0.167
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr7_-_92848837
|
0.166
|
NM_198151
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr22_-_38881529
|
0.163
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr2_+_190540710
|
0.162
|
NM_144708
|
ANKAR
|
ankyrin and armadillo repeat containing
|
|
chr1_-_17766241
|
0.161
|
NM_018715
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr4_+_74606222
|
0.161
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr2_-_175712276
|
0.160
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chrX_+_41548225
|
0.159
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr9_+_34652125
|
0.158
|
NM_001142784
|
IL11RA
|
interleukin 11 receptor, alpha
|
|
chr5_-_135290521
|
0.158
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr4_-_39523132
|
0.156
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr4_+_155484131
|
0.155
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr6_-_138893666
|
0.154
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr22_+_46731297
|
0.152
|
NM_018006
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase
|
|
chr10_-_94257470
|
0.152
|
NM_001165946
|
IDE
|
insulin-degrading enzyme
|
|
chr22_-_19435727
|
0.151
|
NM_001166242
NM_173793
|
C22orf39
|
chromosome 22 open reading frame 39
|
|
chr18_-_47018777
|
0.151
|
|
RPL17
|
ribosomal protein L17
|
|
chr15_+_34261088
|
0.150
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chrX_-_10535476
|
0.150
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr4_-_110723127
|
0.149
|
|
CFI
|
complement factor I
|
|
chr1_+_147400553
|
0.146
|
|
GPR89B
GPR89A
|
G protein-coupled receptor 89B
G protein-coupled receptor 89A
|
|
chr3_+_49506079
|
0.146
|
NM_001177643
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr1_+_174844655
|
0.146
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr5_+_95066628
|
0.145
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr1_+_201617529
|
0.145
|
|
NAV1
|
neuron navigator 1
|
|
chr5_+_42565562
|
0.144
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr6_-_24583454
|
0.144
|
NM_001252328
|
KIAA0319
|
KIAA0319
|
|
chr1_+_151611331
|
0.143
|
|
SNX27
|
sorting nexin family member 27
|
|
chr4_-_110723334
|
0.141
|
NM_000204
|
CFI
|
complement factor I
|
|
chr5_+_140749830
|
0.141
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr3_-_57234279
|
0.140
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr7_+_137761177
|
0.140
|
NM_001190906
NM_001190907
NM_005989
|
AKR1D1
|
aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase)
|
|
chr11_+_12308446
|
0.139
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr3_-_42843217
|
0.139
|
|
|
|
|
chr1_-_109935869
|
0.138
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr3_+_49568370
|
0.137
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr1_-_173020011
|
0.137
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr2_+_102508377
|
0.136
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr7_+_55433140
|
0.136
|
NM_018697
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial)
|
|
chr12_+_48166929
|
0.136
|
NM_017842
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1
|
|
chr5_-_43040147
|
0.136
|
NM_001014279
|
C5orf39
|
chromosome 5 open reading frame 39
|
|
chr1_-_217250230
|
0.135
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr12_+_7023490
|
0.135
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr2_-_202563353
|
0.135
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr16_-_54962707
|
0.131
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr1_-_6420719
|
0.131
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr12_-_16760935
|
0.129
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_157963062
|
0.127
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr5_+_140739589
|
0.125
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr2_-_166060552
|
0.124
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr2_-_151344145
|
0.122
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr12_+_57943843
|
0.122
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr3_-_169587598
|
0.122
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr4_-_17806820
|
0.121
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr10_+_72163914
|
0.121
|
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr19_+_24269877
|
0.121
|
NM_203282
|
ZNF254
|
zinc finger protein 254
|
|
chr12_-_46357949
|
0.120
|
|
|
|
|
chr1_-_149858114
|
0.120
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr4_-_186733372
|
0.120
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr17_+_38975343
|
0.120
|
NM_001195387
NM_145274
NM_001195386
|
TMEM99
|
transmembrane protein 99
|
|
chr13_+_108922560
|
0.119
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr15_-_89456549
|
0.117
|
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
|
chr15_-_89456592
|
0.116
|
NM_001114614
NM_005928
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
|
chr6_-_138820578
|
0.115
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr14_-_60097531
|
0.115
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr17_+_28268622
|
0.114
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|