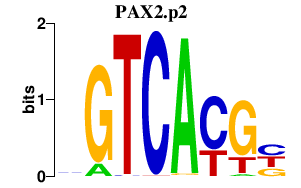
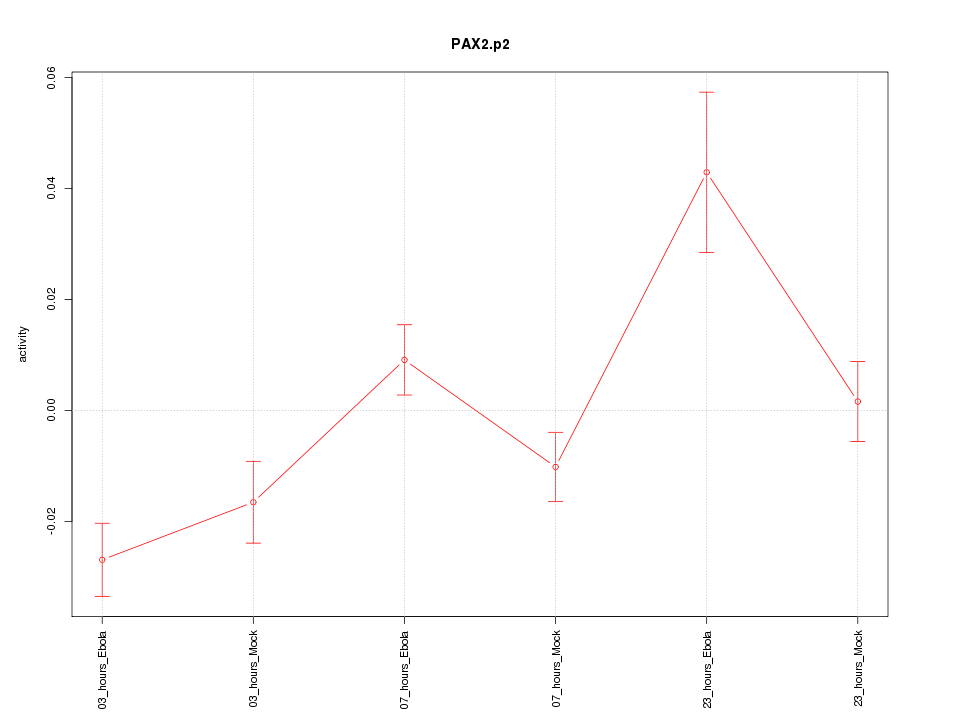
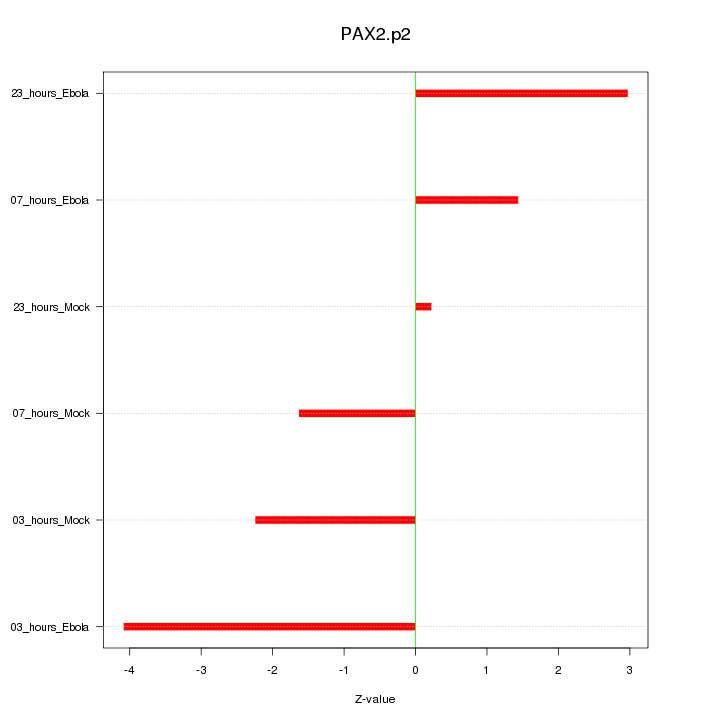
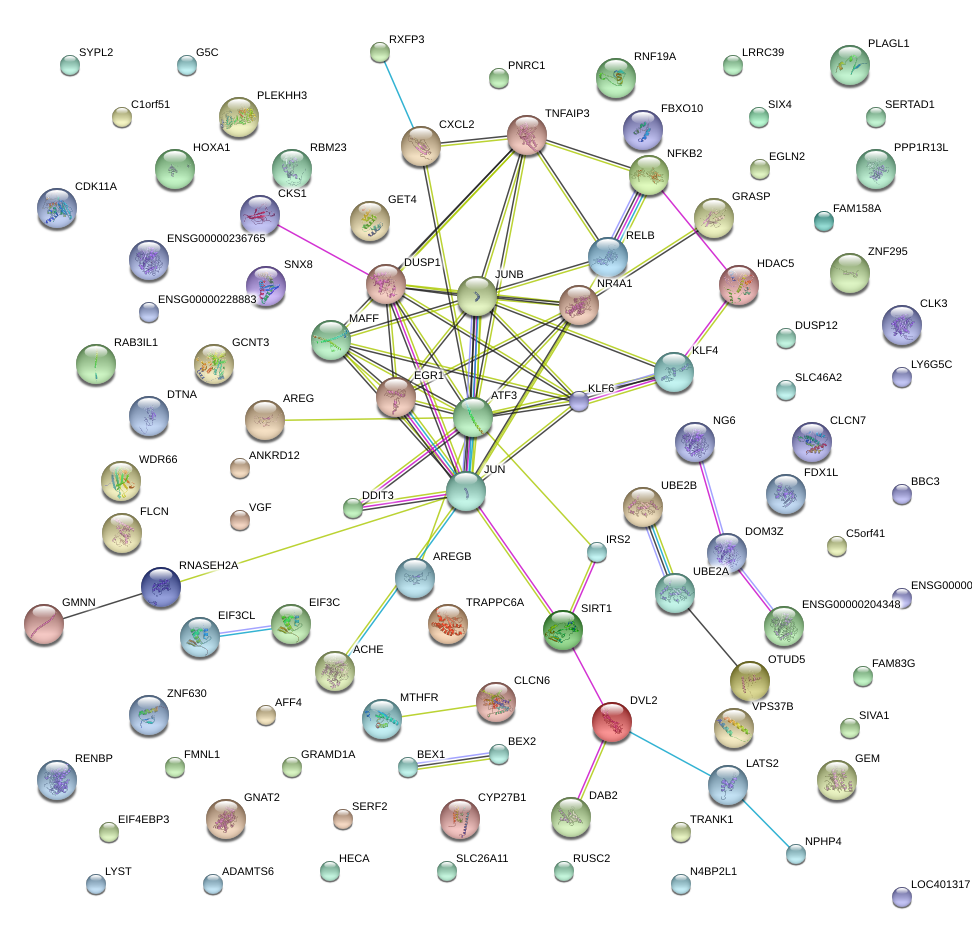
|
chr5_+_172483285
|
3.205
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr13_-_110438896
|
3.132
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr12_+_52445185
|
3.007
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr10_+_104154228
|
2.958
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr10_-_3827205
|
2.804
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr10_-_3827418
|
2.623
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr1_+_11866206
|
2.538
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr19_+_45504686
|
2.529
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr8_-_95274540
|
2.506
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr1_+_11866302
|
2.503
|
|
CLCN6
|
chloride channel 6
|
|
chr22_+_38599026
|
2.411
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr19_-_47734215
|
2.289
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr8_-_95274520
|
2.186
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr1_-_11866079
|
1.965
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr1_+_11866240
|
1.909
|
|
CLCN6
|
chloride channel 6
|
|
chrX_-_102319096
|
1.816
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chr22_+_38598065
|
1.683
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr18_+_9136742
|
1.582
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr17_-_40828441
|
1.575
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr17_-_42201009
|
1.486
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr6_+_89791510
|
1.483
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr17_-_42200957
|
1.464
|
|
HDAC5
|
histone deacetylase 5
|
|
chr4_+_75480628
|
1.424
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr10_+_104155431
|
1.404
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr9_-_115652961
|
1.388
|
NM_033051
|
SLC46A2
|
solute carrier family 46, member 2
|
|
chr19_-_45908288
|
1.388
|
NM_006663
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr1_+_212781969
|
1.385
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr18_+_32398225
|
1.352
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr12_-_105501410
|
1.349
|
|
LOC414300
|
uncharacterized LOC414300
|
|
chr11_-_61684981
|
1.316
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr5_+_137801178
|
1.287
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr21_-_43430448
|
1.256
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr12_-_58159386
|
1.249
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr1_-_100643770
|
1.238
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr9_-_110252045
|
1.212
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr7_+_28448970
|
1.200
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr1_+_212782103
|
1.200
|
|
ATF3
|
activating transcription factor 3
|
|
chr19_-_45681454
|
1.198
|
NM_024108
|
TRAPPC6A
|
trafficking protein particle complex 6A
|
|
chr1_+_150255228
|
1.167
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chrX_-_48814758
|
1.164
|
|
OTUD5
|
OTU domain containing 5
|
|
chr1_+_110009099
|
1.161
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr7_-_100808832
|
1.161
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr7_+_916240
|
1.155
|
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr10_+_69644341
|
1.141
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr6_+_89790464
|
1.138
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr12_+_52400725
|
1.133
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr9_-_110251754
|
1.132
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr17_+_43299277
|
1.129
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr5_+_33936490
|
1.107
|
NM_016568
|
RXFP3
|
relaxin/insulin-like family peptide receptor 3
|
|
chr1_-_59248287
|
1.103
|
|
JUN
|
jun proto-oncogene
|
|
chr5_-_132299263
|
1.093
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr14_-_23388380
|
1.091
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr12_+_122356446
|
1.090
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr5_-_172198160
|
1.074
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr14_+_105219516
|
1.062
|
|
SIVA1
|
SIVA1, apoptosis-inducing factor
|
|
chr6_+_138188352
|
1.052
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr6_+_89790412
|
1.044
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr16_-_1525020
|
1.030
|
|
CLCN7
|
chloride channel 7
|
|
chrX_-_153210057
|
1.028
|
NM_002910
|
RENBP
|
renin binding protein
|
|
chr19_+_12902295
|
1.026
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr13_-_21635629
|
1.021
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr6_+_139456240
|
1.018
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr5_+_137801166
|
1.007
|
|
EGR1
|
early growth response 1
|
|
chr19_-_10426673
|
1.006
|
NM_001031734
|
FDX1L
|
ferredoxin 1-like
|
|
chr3_+_194207685
|
1.002
|
|
FLJ34208
|
uncharacterized LOC401106
|
|
chr9_-_37576248
|
1.000
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr5_-_132299248
|
0.990
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr5_+_139927182
|
0.979
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr3_-_36986535
|
0.979
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr16_-_1525006
|
0.978
|
|
CLCN7
|
chloride channel 7
|
|
chr6_-_144329254
|
0.971
|
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr1_-_236030219
|
0.969
|
NM_000081
|
LYST
|
lysosomal trafficking regulator
|
|
chr17_-_18907988
|
0.955
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr7_-_2354010
|
0.955
|
|
SNX8
|
sorting nexin 8
|
|
chr15_+_74908195
|
0.936
|
|
CLK3
|
CDC-like kinase 3
|
|
chr7_+_916178
|
0.933
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr1_-_6052481
|
0.933
|
NM_015102
|
NPHP4
|
nephronophthisis 4
|
|
chr1_-_11865963
|
0.932
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr4_-_74964836
|
0.929
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chrX_-_102319080
|
0.928
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr17_-_7137675
|
0.916
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr5_+_133706869
|
0.916
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr6_-_31648140
|
0.910
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr9_+_35490018
|
0.909
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr14_-_23388294
|
0.907
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr1_-_1590403
|
0.902
|
|
CDK11A
CDK11B
|
cyclin-dependent kinase 11A
cyclin-dependent kinase 11B
|
|
chr1_-_236030170
|
0.896
|
|
|
|
|
chr17_-_17140425
|
0.892
|
|
FLCN
|
folliculin
|
|
chr5_-_64777703
|
0.889
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr19_+_12902286
|
0.871
|
|
|
|
|
chr12_-_123380608
|
0.870
|
|
|
|
|
chrX_-_48815088
|
0.869
|
|
OTUD5
|
OTU domain containing 5
|
|
chr19_-_40931880
|
0.864
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr15_+_59908908
|
0.861
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chrX_-_47931000
|
0.856
|
NM_001190255
|
ZNF630
|
zinc finger protein 630
|
|
chr5_-_39424930
|
0.846
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr8_-_101322180
|
0.843
|
|
RNF19A
|
ring finger protein 19A
|
|
chr19_+_35491225
|
0.842
|
NM_001136199
NM_020895
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr14_-_24610741
|
0.834
|
NM_016049
|
FAM158A
|
family with sequence similarity 158, member A
|
|
chr16_-_28415151
|
0.834
|
NM_001099661
NM_001037808
NM_003752
|
EIF3CL
EIF3C
|
eukaryotic translation initiation factor 3, subunit C-like
eukaryotic translation initiation factor 3, subunit C
|
|
chr15_+_59908724
|
0.833
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr15_+_44084039
|
0.831
|
NM_001018108
NM_001199875
NM_001199876
|
SERF2
|
small EDRK-rich factor 2
|
|
chr22_+_38597938
|
0.823
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chrX_-_48814853
|
0.822
|
NM_001136157
NM_001136158
NM_017602
|
OTUD5
|
OTU domain containing 5
|
|
chr7_-_2354050
|
0.820
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr9_+_35489989
|
0.818
|
NM_014806
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr16_-_1524926
|
0.815
|
|
CLCN7
|
chloride channel 7
|
|
chr13_-_33002218
|
0.814
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr17_-_7137580
|
0.810
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr1_-_110155672
|
0.796
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr7_-_27135524
|
0.788
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr12_-_57914287
|
0.786
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr12_-_123380628
|
0.786
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr17_+_78194090
|
0.785
|
NM_001166347
NM_001166348
NM_001166349
NM_173626
|
SLC26A11
|
solute carrier family 26, member 11
|
|
chr7_-_100493481
|
0.784
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr19_+_41305041
|
0.780
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr14_-_61191027
|
0.780
|
|
SIX4
|
SIX homeobox 4
|
|
chr17_+_41003360
|
0.775
|
|
|
|
|
chr6_-_31939658
|
0.772
|
|
DOM3Z
|
dom-3 homolog Z (C. elegans)
|
|
chr15_-_72410226
|
0.771
|
NM_006901
|
MYO9A
|
myosin IXA
|
|
chr21_+_30517897
|
0.767
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr18_+_60382617
|
0.764
|
NM_194449
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr19_+_41305100
|
0.759
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr8_-_29207795
|
0.759
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr3_-_53925871
|
0.759
|
NM_021237
|
SELK
|
selenoprotein K
|
|
chr22_+_40573879
|
0.757
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr8_-_103667882
|
0.754
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chrX_+_70364680
|
0.754
|
NM_001166660
NM_018977
NM_181303
|
NLGN3
|
neuroligin 3
|
|
chr4_-_74964996
|
0.752
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr1_-_6052410
|
0.752
|
|
NPHP4
|
nephronophthisis 4
|
|
chr19_+_36545804
|
0.752
|
|
WDR62
|
WD repeat domain 62
|
|
chr1_-_201438150
|
0.741
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr1_+_113498984
|
0.741
|
|
LOC100506392
|
uncharacterized LOC100506392
|
|
chr19_+_35491329
|
0.737
|
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr3_-_169899519
|
0.736
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr7_-_6312137
|
0.729
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr16_-_28414940
|
0.724
|
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr17_-_40828078
|
0.721
|
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr5_+_149380168
|
0.717
|
NM_014983
|
HMGXB3
|
HMG box domain containing 3
|
|
chr22_-_39268169
|
0.715
|
|
CBX6
|
chromobox homolog 6
|
|
chr2_-_220042731
|
0.713
|
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr7_+_27135983
|
0.712
|
|
|
|
|
chr3_+_19988530
|
0.708
|
NM_004162
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr19_+_1067293
|
0.706
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr6_-_30712294
|
0.703
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr9_+_74526369
|
0.701
|
NM_182505
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr1_+_22778343
|
0.698
|
NM_001083621
NM_014870
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr14_-_20929484
|
0.696
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr17_-_7137722
|
0.693
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr9_-_22009270
|
0.693
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr9_-_99382073
|
0.692
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr17_-_63052884
|
0.691
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr17_+_40688264
|
0.691
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr22_+_42229268
|
0.690
|
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr12_+_72057676
|
0.684
|
NM_031435
|
THAP2
|
THAP domain containing, apoptosis associated protein 2
|
|
chr20_+_61867234
|
0.684
|
NM_022161
NM_139317
|
BIRC7
|
baculoviral IAP repeat containing 7
|
|
chr4_+_72897520
|
0.682
|
NM_004885
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chr1_-_193155723
|
0.681
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr10_+_69644708
|
0.681
|
|
SIRT1
|
sirtuin 1
|
|
chr22_-_39268230
|
0.679
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr7_-_36429695
|
0.679
|
NM_001100425
|
KIAA0895
|
KIAA0895
|
|
chr19_-_460995
|
0.674
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr19_+_18118921
|
0.671
|
NM_015683
|
ARRDC2
|
arrestin domain containing 2
|
|
chr17_+_18218605
|
0.667
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr20_-_60927394
|
0.664
|
|
LAMA5
|
laminin, alpha 5
|
|
chr20_+_51589752
|
0.662
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr15_-_72410108
|
0.662
|
|
MYO9A
|
myosin IXA
|
|
chr5_-_1112106
|
0.662
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr22_-_24059436
|
0.662
|
|
|
|
|
chr11_+_108093558
|
0.660
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr17_-_62833264
|
0.658
|
|
PLEKHM1P
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene
|
|
chr17_+_36507708
|
0.655
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr17_+_18218569
|
0.654
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr19_+_36545757
|
0.653
|
NM_001083961
NM_173636
|
WDR62
|
WD repeat domain 62
|
|
chr17_+_28443822
|
0.652
|
NM_032141
|
NSRP1
|
nuclear speckle splicing regulatory protein 1
|
|
chr2_-_20850822
|
0.647
|
NM_022460
|
HS1BP3
|
HCLS1 binding protein 3
|
|
chr19_-_36545646
|
0.646
|
NM_152658
|
THAP8
|
THAP domain containing 8
|
|
chrX_-_48937560
|
0.645
|
NM_001029896
|
WDR45
|
WD repeat domain 45
|
|
chr9_+_74526527
|
0.645
|
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr6_-_31745048
|
0.644
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr5_-_132298973
|
0.643
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr15_+_90544608
|
0.642
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr6_-_53530505
|
0.642
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr10_+_134210691
|
0.640
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr6_-_144329342
|
0.639
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr5_-_132299191
|
0.638
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr19_-_4867655
|
0.634
|
|
PLIN3
|
perilipin 3
|
|
chr14_-_68283293
|
0.631
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr22_-_39268214
|
0.630
|
|
CBX6
|
chromobox homolog 6
|
|
chr1_-_935352
|
0.627
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr1_-_39325317
|
0.625
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr17_+_40688189
|
0.625
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr17_-_62833246
|
0.625
|
|
PLEKHM1P
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene
|
|
chr19_-_4867638
|
0.624
|
|
PLIN3
|
perilipin 3
|
|
chr17_-_79885544
|
0.622
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr13_+_48877775
|
0.620
|
NM_000321
|
RB1
|
retinoblastoma 1
|
|
chr1_+_44440642
|
0.619
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_-_79881304
|
0.619
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr18_+_29078026
|
0.618
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr1_-_38273823
|
0.618
|
|
YRDC
|
yrdC domain containing (E. coli)
|