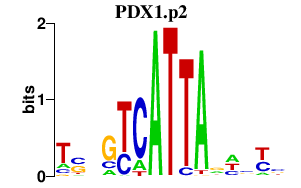
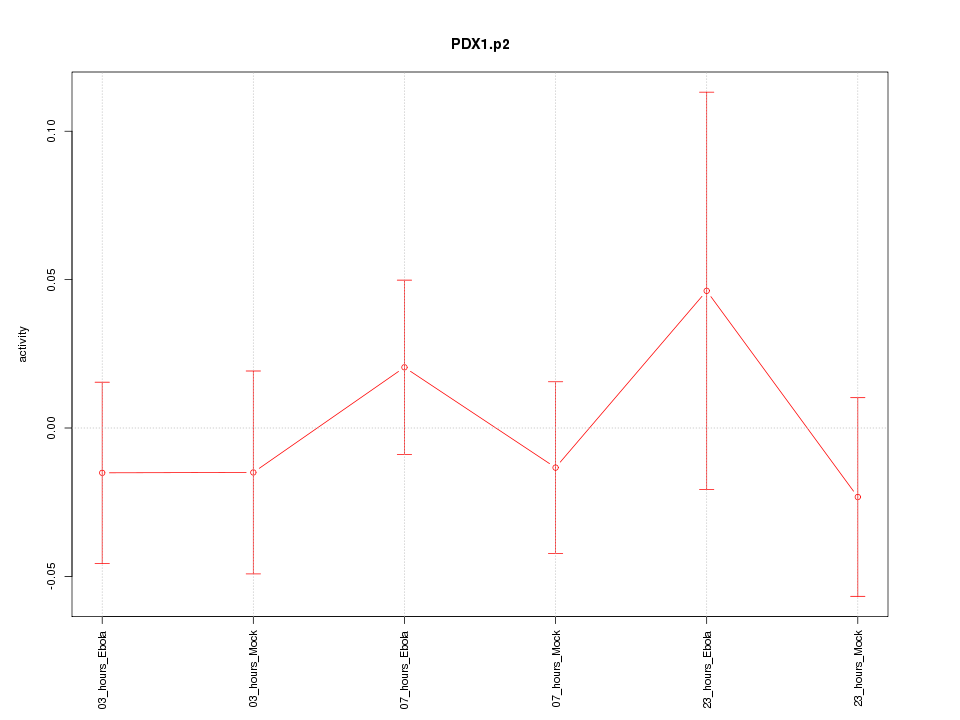
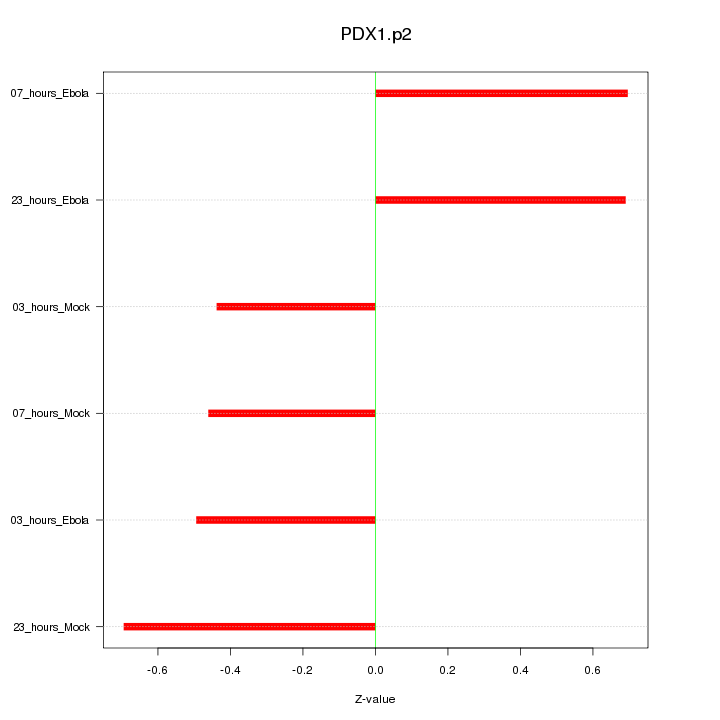
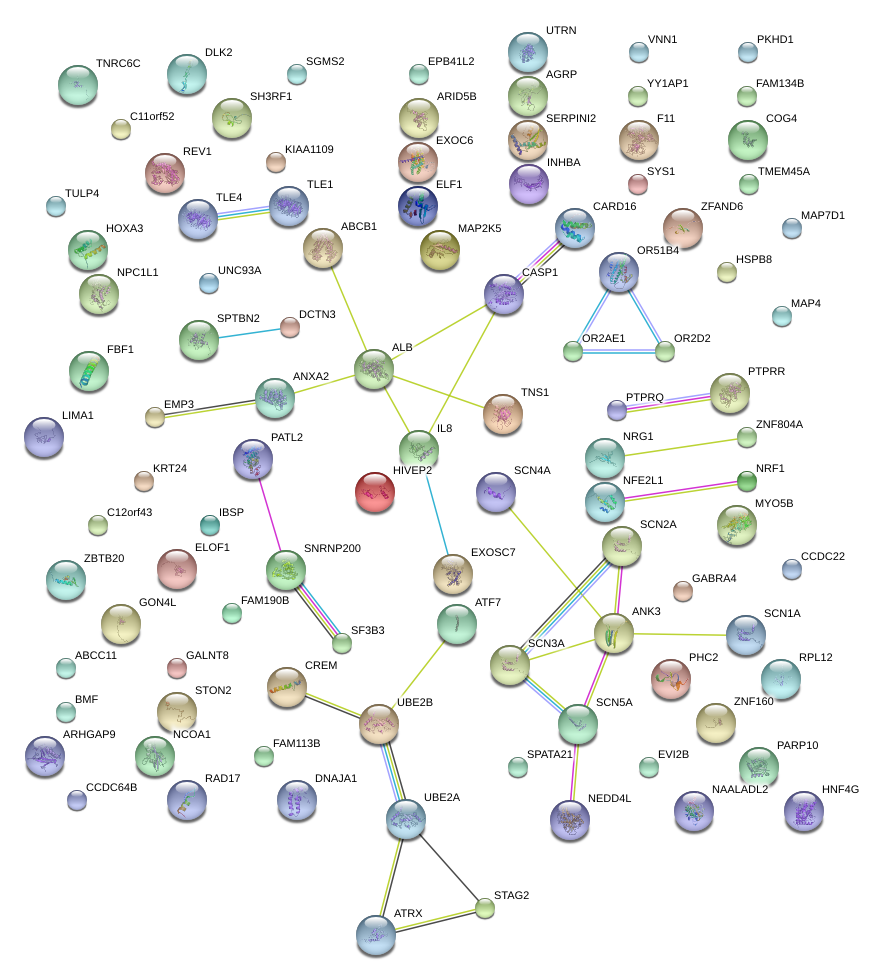
|
chr4_+_108814419
|
1.105
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr8_+_32579350
|
0.724
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr4_+_74606222
|
0.668
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr15_-_44969085
|
0.625
|
NM_001145112
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast)
|
|
chr12_-_54019633
|
0.593
|
NM_001206683
|
ATF7
|
activating transcription factor 7
|
|
chr6_-_43423785
|
0.592
|
NM_206539
|
DLK2
|
delta-like 2 homolog (Drosophila)
|
|
chr10_+_63808968
|
0.491
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr17_+_47923271
|
0.455
|
NM_001242791
|
FLJ45513
|
uncharacterized LOC729220
|
|
chr8_+_76452202
|
0.428
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr6_-_51952368
|
0.423
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr18_+_55888750
|
0.403
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55888799
|
0.394
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr15_+_67841341
|
0.392
|
NM_001206804
|
MAP2K5
|
mitogen-activated protein kinase kinase 5
|
|
chr12_+_47610244
|
0.378
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr16_-_70557430
|
0.346
|
NM_001195139
NM_015386
|
COG4
|
component of oligomeric golgi complex 4
|
|
chr16_-_3085541
|
0.345
|
NM_001103175
|
CCDC64B
|
coiled-coil domain containing 64B
|
|
chr3_-_167191808
|
0.326
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr14_-_81864726
|
0.322
|
NM_033104
|
STON2
|
stonin 2
|
|
chr12_+_119616594
|
0.295
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr1_+_36636690
|
0.285
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr15_-_40398286
|
0.282
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr6_-_143266283
|
0.255
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr2_-_166060552
|
0.253
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr16_-_67517715
|
0.221
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr18_+_55888838
|
0.221
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr4_+_88720701
|
0.214
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr1_-_33841142
|
0.214
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr6_-_133035180
|
0.210
|
NM_004666
|
VNN1
|
vanin 1
|
|
chrX_+_49091926
|
0.209
|
NM_014008
|
CCDC22
|
coiled-coil domain containing 22
|
|
chr3_+_174577069
|
0.198
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr3_+_100238063
|
0.195
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr3_-_114343791
|
0.187
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_-_5323174
|
0.175
|
NM_033179
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4
|
|
chr6_+_167704802
|
0.167
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr3_-_114478117
|
0.164
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_121454285
|
0.164
|
NM_022895
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr10_-_62332666
|
0.156
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chrX_+_49091931
|
0.154
|
|
CCDC22
|
coiled-coil domain containing 22
|
|
chr19_-_36019194
|
0.148
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr9_+_33026482
|
0.148
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr13_-_41593491
|
0.145
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr12_-_54020081
|
0.141
|
NM_001130060
NM_001206682
NM_006856
|
ATF7
|
activating transcription factor 7
|
|
chr12_-_57873364
|
0.140
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr8_-_145060594
|
0.139
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr14_-_77889231
|
0.127
|
NM_001113475
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1
|
|
chr11_-_104905849
|
0.127
|
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr2_+_185463235
|
0.124
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr20_+_43990576
|
0.118
|
NM_001197129
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr7_-_44580857
|
0.117
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr18_-_47376156
|
0.115
|
|
MYO5B
|
myosin VB
|
|
chr7_+_116395440
|
0.111
|
|
|
|
|
chr4_+_123161119
|
0.111
|
|
KIAA1109
|
KIAA1109
|
|
chr16_+_70557688
|
0.108
|
NM_012426
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr4_+_187187342
|
0.106
|
|
F11
|
coagulation factor XI
|
|
chr7_-_41742666
|
0.103
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr16_-_48269087
|
0.097
|
NM_032583
NM_145186
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr17_+_76000248
|
0.094
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr15_-_40398573
|
0.093
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr19_+_48828627
|
0.085
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr10_+_35484829
|
0.082
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr6_+_144612872
|
0.079
|
NM_007124
|
UTRN
|
utrophin
|
|
chr7_-_87342569
|
0.076
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr7_-_99474655
|
0.069
|
NM_001005276
|
OR2AE1
|
olfactory receptor, family 2, subfamily AE, member 1
|
|
chr12_+_4829751
|
0.068
|
NM_017417
|
GALNT8
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8)
|
|
chr6_+_158733691
|
0.067
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chrX_-_76814217
|
0.067
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr4_-_170077807
|
0.066
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr11_+_111789600
|
0.065
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr9_+_82187599
|
0.065
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr9_+_82187469
|
0.064
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr5_-_16509106
|
0.062
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr12_+_80838125
|
0.061
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr2_+_24807345
|
0.060
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr17_-_38859976
|
0.058
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr2_+_204055404
|
0.054
|
|
RPL12
|
ribosomal protein L12
|
|
chr5_+_68667280
|
0.053
|
NM_133339
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr4_-_46996423
|
0.051
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr16_+_70557726
|
0.050
|
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr3_+_45017729
|
0.050
|
NM_015004
|
EXOSC7
|
exosome component 7
|
|
chr5_-_172083569
|
0.049
|
|
|
|
|
chr12_-_50616425
|
0.047
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr9_+_33624214
|
0.045
|
|
ANXA2P2
|
annexin A2 pseudogene 2
|
|
chrX_+_123234462
|
0.045
|
|
STAG2
|
stromal antigen 2
|
|
chr15_+_80364902
|
0.045
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr1_-_155649597
|
0.045
|
NM_001198899
|
YY1AP1
|
YY1 associated protein 1
|
|
chr11_-_6913730
|
0.045
|
NM_003700
|
OR2D2
|
olfactory receptor, family 2, subfamily D, member 2
|
|
chr7_+_129269916
|
0.044
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr3_-_47933997
|
0.044
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr7_-_27166638
|
0.042
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr17_-_29641094
|
0.042
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_-_155649243
|
0.041
|
|
|
|
|
chrX_+_118708540
|
0.040
|
|
UBE2A
|
ubiquitin-conjugating enzyme E2A
|
|
chr10_+_94594469
|
0.040
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr4_+_74281884
|
0.039
|
|
ALB
|
albumin
|
|
chr2_-_100046262
|
0.037
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr6_-_131291480
|
0.037
|
NM_001199389
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr2_-_218808705
|
0.035
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr11_-_66488712
|
0.034
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr2_-_96947640
|
0.033
|
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5)
|
|
chr1_-_16763887
|
0.033
|
NM_198546
|
SPATA21
|
spermatogenesis associated 21
|
|
chr10_+_86184685
|
0.033
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr9_-_34620483
|
0.032
|
|
DCTN3
|
dynactin 3 (p22)
|
|
chr2_+_185463292
|
0.031
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr2_-_216259197
|
0.028
|
|
FN1
|
fibronectin 1
|
|
chr11_+_22696318
|
0.027
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr4_+_77356252
|
0.024
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr14_+_101295409
|
0.023
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chrX_-_132095422
|
0.022
|
NM_001077188
NM_147175
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr5_+_140227356
|
0.022
|
NM_014005
NM_031857
|
PCDHA9
|
protocadherin alpha 9
|
|
chr1_+_17906941
|
0.020
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr17_-_7082654
|
0.020
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr16_+_29674282
|
0.019
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr3_+_11178778
|
0.018
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr17_-_29641068
|
0.018
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr14_+_23789396
|
0.017
|
NM_004643
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr4_-_24981769
|
0.016
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr17_-_42143937
|
0.016
|
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr6_+_25754926
|
0.015
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chr6_+_131571298
|
0.015
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr12_-_121454210
|
0.014
|
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr3_+_45017749
|
0.014
|
|
EXOSC7
|
exosome component 7
|
|
chr16_+_29674979
|
0.014
|
|
SPN
|
sialophorin
|
|
chr5_-_142815076
|
0.013
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr7_+_104681378
|
0.012
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chrX_-_62780748
|
0.012
|
|
LOC92249
|
uncharacterized LOC92249
|
|
chr14_-_23504353
|
0.011
|
NM_001144932
NM_002797
NM_001130725
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr12_+_105153258
|
0.011
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr1_+_79086087
|
0.010
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr6_+_147524866
|
0.010
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr17_+_68071386
|
0.007
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chrX_-_118726852
|
0.007
|
NM_001173488
|
NKRF
|
NFKB repressing factor
|
|
chr18_+_28898051
|
0.006
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr17_-_29641087
|
0.006
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr4_+_123300120
|
0.005
|
NM_139243
|
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific)
|
|
chr1_+_28764660
|
0.004
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr2_+_228192227
|
0.004
|
NM_020194
|
MFF
|
mitochondrial fission factor
|
|
chr4_-_72649734
|
0.003
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr12_+_15475190
|
0.002
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr17_-_1412995
|
0.002
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr16_-_28620648
|
0.001
|
NM_177530
NM_177534
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr3_+_52811601
|
0.000
|
NM_002215
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr6_-_131321868
|
0.000
|
NM_001252660
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|