|
chr19_-_50143350
|
7.150
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr7_+_18535351
|
5.460
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr1_+_68150858
|
5.157
|
NM_001199741
NM_001199742
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr1_+_68151092
|
5.034
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr6_-_11232883
|
4.253
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_11232901
|
4.117
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_11232900
|
3.836
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr7_-_100183722
|
3.431
|
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr6_-_53013581
|
3.244
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr22_-_24096551
|
3.077
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr16_-_75285383
|
3.033
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr6_-_143266283
|
2.976
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr7_+_112120907
|
2.684
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr19_+_41305041
|
2.497
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_+_41305100
|
2.426
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr7_-_141673572
|
2.417
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr6_+_26183957
|
2.412
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr1_+_201979689
|
2.391
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr19_-_40931880
|
2.359
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr1_+_201979747
|
2.191
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr11_+_537521
|
2.090
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr7_-_100183753
|
1.965
|
NM_002319
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr19_-_1132109
|
1.932
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr17_-_8063946
|
1.913
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr13_+_52027435
|
1.782
|
|
|
|
|
chr6_+_20402040
|
1.732
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr10_-_64578926
|
1.724
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr18_+_32290195
|
1.719
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr19_-_893184
|
1.709
|
|
MED16
|
mediator complex subunit 16
|
|
chr12_-_11150473
|
1.700
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr6_+_31588460
|
1.677
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr13_+_97794050
|
1.667
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr19_+_41305333
|
1.641
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_-_3971089
|
1.620
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr11_+_112047088
|
1.615
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr19_-_18392048
|
1.544
|
|
JUND
|
jun D proto-oncogene
|
|
chr6_+_37137882
|
1.494
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr3_+_119316694
|
1.485
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr1_+_228645807
|
1.473
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr12_+_7053180
|
1.470
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr17_-_7832752
|
1.433
|
NM_004732
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3
|
|
chr3_-_87040268
|
1.428
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr15_+_78384926
|
1.418
|
NM_001101404
|
SH2D7
|
SH2 domain containing 7
|
|
chr6_+_31588444
|
1.411
|
NM_004638
NM_080686
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr11_+_64073040
|
1.404
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr6_+_31588524
|
1.400
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr2_-_211035913
|
1.391
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr19_-_893215
|
1.341
|
NM_005481
|
MED16
|
mediator complex subunit 16
|
|
chr17_+_72983723
|
1.336
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr14_+_23352430
|
1.335
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr1_-_196577438
|
1.332
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr19_-_18392413
|
1.329
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr5_+_60628085
|
1.316
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr6_+_31588523
|
1.307
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr19_-_55919324
|
1.304
|
NM_014501
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr9_-_3223007
|
1.303
|
|
|
|
|
chr12_-_10978867
|
1.293
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chr19_-_1021122
|
1.278
|
NM_001033026
NM_033420
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr1_-_154832187
|
1.262
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr16_-_30107492
|
1.253
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr6_-_128222175
|
1.250
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr11_+_64085559
|
1.175
|
NM_012094
NM_181651
NM_181652
|
PRDX5
|
peroxiredoxin 5
|
|
chr22_+_38092994
|
1.160
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr6_+_43139190
|
1.139
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr16_+_30107742
|
1.130
|
|
|
|
|
chr2_-_207082626
|
1.127
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr1_-_6052481
|
1.121
|
NM_015102
|
NPHP4
|
nephronophthisis 4
|
|
chr4_+_184020343
|
1.108
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr19_-_18392436
|
1.091
|
|
JUND
|
jun D proto-oncogene
|
|
chr1_-_94586678
|
1.081
|
NM_000350
|
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr12_-_11091805
|
1.077
|
NM_023922
|
TAS2R14
|
taste receptor, type 2, member 14
|
|
chr14_-_23445885
|
1.066
|
|
AJUBA
|
ajuba LIM protein
|
|
chr14_-_68283293
|
1.060
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr11_-_64084980
|
1.058
|
NM_016404
|
TRMT112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae)
|
|
chr1_-_196577676
|
1.035
|
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr1_-_2457988
|
1.032
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr18_+_44526786
|
0.994
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr11_+_64085602
|
0.975
|
|
PRDX5
|
peroxiredoxin 5
|
|
chr3_-_87040246
|
0.975
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
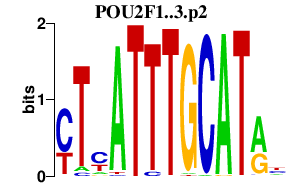
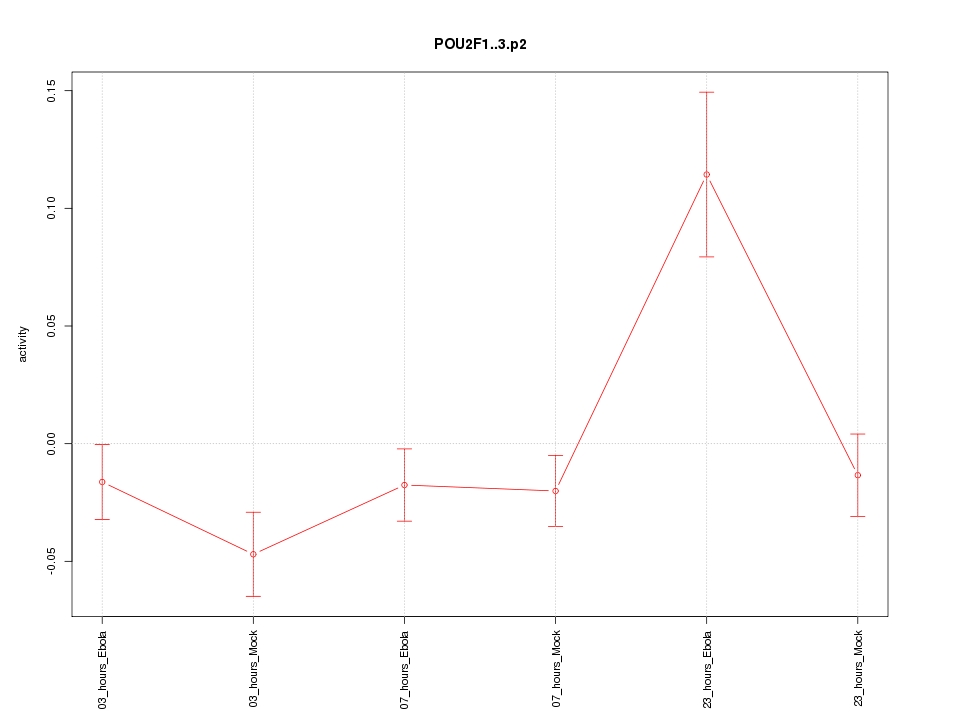
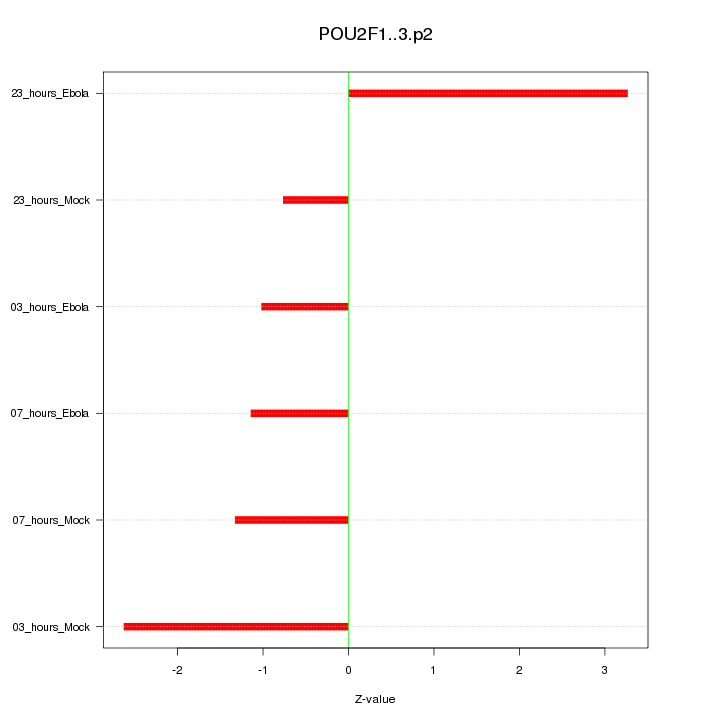
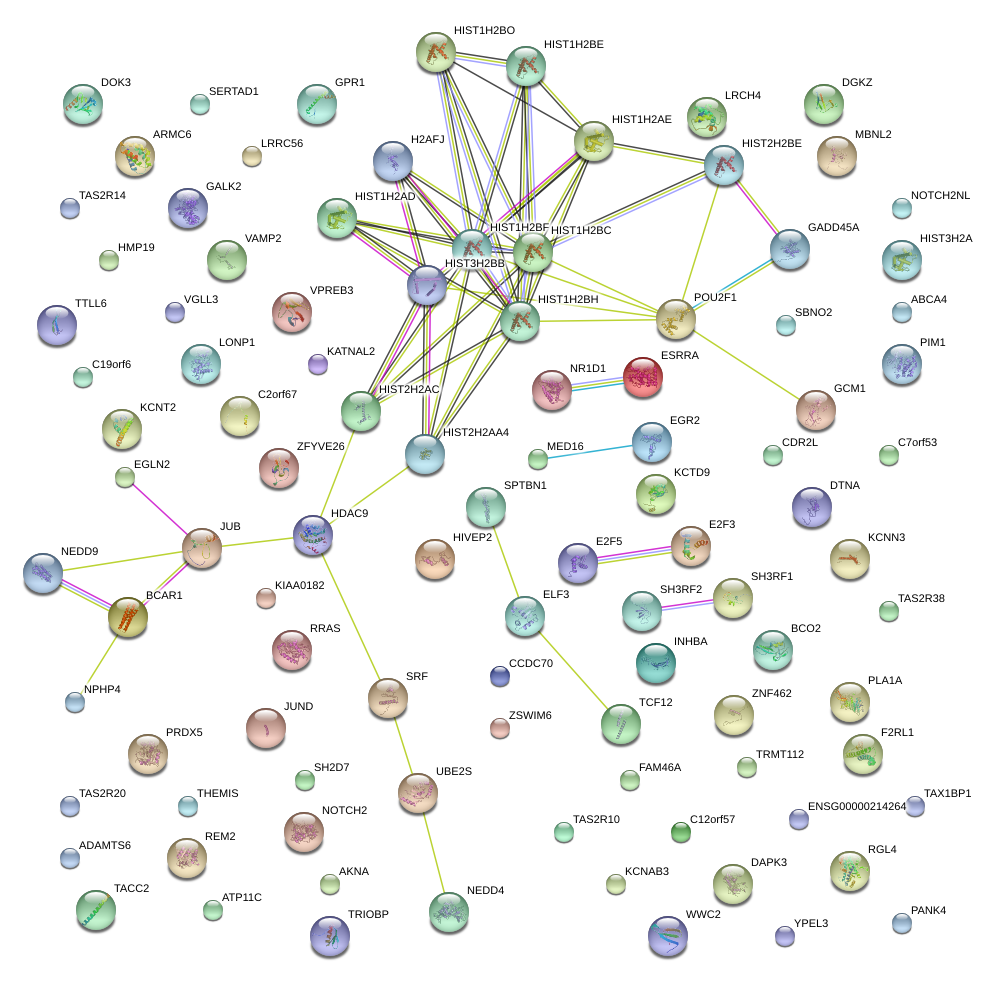
chr1_+_167298280
|
0.975
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr5_+_145317297
|
0.970
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr5_-_176935877
|
0.966
|
|
DOK3
|
docking protein 3
|
|
chr17_-_38256548
|
0.961
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr7_-_41742666
|
0.940
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr15_+_57511616
|
0.927
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr1_-_6052410
|
0.918
|
|
NPHP4
|
nephronophthisis 4
|
|
chr16_+_85645014
|
0.918
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr5_+_173472692
|
0.917
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chrX_-_138914384
|
0.910
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr22_+_24033047
|
0.903
|
NM_153615
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4
|
|
chr8_-_25315941
|
0.902
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr1_-_228645517
|
0.889
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr1_+_149858466
|
0.877
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr5_+_76114832
|
0.870
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr5_-_64777703
|
0.867
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr1_-_120484230
|
0.855
|
|
NOTCH2
|
notch 2
|
|
chr10_+_123923104
|
0.830
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_+_46383115
|
0.828
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr19_+_19144617
|
0.818
|
|
ARMC6
|
armadillo repeat containing 6
|
|
chr7_+_27778991
|
0.818
|
NM_001206901
|
TAX1BP1
|
Tax1 (human T-cell leukemia virus type I) binding protein 1
|
|
chr9_-_117160748
|
0.813
|
|
AKNA
|
AT-hook transcription factor
|
|
chr2_+_54785453
|
0.799
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr6_-_82462374
|
0.797
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr6_+_43139201
|
0.790
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr15_-_56209305
|
0.785
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr9_+_109686436
|
0.778
|
|
ZNF462
|
zinc finger protein 462
|
|
chr19_+_19144370
|
0.759
|
NM_001199196
NM_033415
|
ARMC6
|
armadillo repeat containing 6
|
|
chr6_+_43138919
|
0.755
|
NM_003131
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr4_-_170077807
|
0.745
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr13_+_52436116
|
0.740
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr15_+_49462408
|
0.736
|
NM_002044
|
GALK2
|
galactokinase 2
|
|
chr17_-_46894380
|
0.726
|
NM_001130918
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr8_+_86099909
|
0.725
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr7_+_100183926
|
0.722
|
NM_033506
|
FBXO24
|
F-box protein 24
|
|
chr1_+_167298382
|
0.715
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr19_-_1020995
|
0.713
|
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr5_-_141030934
|
0.700
|
|
FCHSD1
|
FCH and double SH3 domains 1
|
|
chrX_-_48776282
|
0.700
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr19_-_19144326
|
0.699
|
NM_001017392
NM_014884
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr10_+_123922940
|
0.691
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr4_+_74286846
|
0.690
|
|
ALB
|
albumin
|
|
chr19_-_19144304
|
0.686
|
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr2_-_97405775
|
0.666
|
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr7_+_141490016
|
0.662
|
NM_018980
|
TAS2R5
|
taste receptor, type 2, member 5
|
|
chr6_-_33679435
|
0.660
|
|
C6orf125
|
chromosome 6 open reading frame 125
|
|
chr7_-_44122105
|
0.655
|
NM_013284
|
POLM
|
polymerase (DNA directed), mu
|
|
chr19_+_19144520
|
0.648
|
|
ARMC6
|
armadillo repeat containing 6
|
|
chr19_-_19144262
|
0.647
|
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr6_-_43655514
|
0.644
|
|
MRPS18A
|
mitochondrial ribosomal protein S18A
|
|
chr2_-_97405799
|
0.640
|
NM_001142292
NM_030805
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr7_-_32982574
|
0.639
|
|
RP9P
|
retinitis pigmentosa 9 pseudogene
|
|
chr8_-_25315791
|
0.633
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr12_-_57443858
|
0.632
|
NM_005379
|
MYO1A
|
myosin IA
|
|
chr12_+_14518608
|
0.624
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr10_-_64576074
|
0.614
|
|
EGR2
|
early growth response 2
|
|
chrX_+_117526665
|
0.609
|
|
WDR44
|
WD repeat domain 44
|
|
chr17_-_73127787
|
0.607
|
NM_001252377
NM_014595
|
NT5C
|
5', 3'-nucleotidase, cytosolic
|
|
chr4_+_72897520
|
0.597
|
NM_004885
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chr19_-_50168866
|
0.593
|
|
IRF3
|
interferon regulatory factor 3
|
|
chr6_+_135516879
|
0.589
|
|
|
|
|
chr18_+_32073245
|
0.589
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr19_+_35531409
|
0.585
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr17_+_38498270
|
0.582
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr12_+_122064448
|
0.581
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr6_-_43655539
|
0.579
|
NM_001193343
NM_018135
|
MRPS18A
|
mitochondrial ribosomal protein S18A
|
|
chr10_-_64576123
|
0.571
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr22_-_43010942
|
0.565
|
NM_032311
NM_178136
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr1_-_193155723
|
0.563
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr21_+_30452872
|
0.557
|
NM_020152
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr7_-_44122036
|
0.554
|
|
POLM
|
polymerase (DNA directed), mu
|
|
chr12_-_31479084
|
0.545
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr12_-_31479120
|
0.541
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chrX_+_123234462
|
0.539
|
|
STAG2
|
stromal antigen 2
|
|
chr3_-_192445344
|
0.531
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr21_-_11098819
|
0.521
|
NM_001187
NM_182484
NM_181704
NM_182481
NM_182482
|
BAGE
BAGE5
BAGE4
BAGE3
BAGE2
|
B melanoma antigen
B melanoma antigen family, member 5
B melanoma antigen family, member 4
B melanoma antigen family, member 3
B melanoma antigen family, member 2
|
|
chr9_-_32526164
|
0.515
|
NM_014314
|
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58
|
|
chr10_-_62332666
|
0.507
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_170632274
|
0.506
|
|
PRRX1
|
paired related homeobox 1
|
|
chr15_-_41364176
|
0.505
|
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr3_+_50192420
|
0.502
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr9_+_136325086
|
0.501
|
NM_001135775
NM_001242369
NM_001242370
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr5_+_173472737
|
0.488
|
|
HMP19
|
HMP19 protein
|
|
chr2_+_86668270
|
0.478
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr12_+_53442732
|
0.473
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr5_-_139682680
|
0.472
|
NM_002622
|
PFDN1
|
prefoldin subunit 1
|
|
chr12_-_50616425
|
0.468
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr5_-_139682674
|
0.462
|
|
PFDN1
|
prefoldin subunit 1
|
|
chr5_-_175964824
|
0.459
|
|
RNF44
|
ring finger protein 44
|
|
chr12_-_31479038
|
0.459
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr6_-_35656677
|
0.451
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr22_-_43010837
|
0.450
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr11_-_34379526
|
0.449
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr1_-_149859465
|
0.448
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr5_-_141030978
|
0.442
|
NM_033449
|
FCHSD1
|
FCH and double SH3 domains 1
|
|
chr4_-_74088773
|
0.439
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr19_+_17378148
|
0.438
|
NM_001033549
NM_014173
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr7_-_112727832
|
0.438
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr12_-_86230158
|
0.437
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr14_-_23540770
|
0.436
|
NM_001164816
NM_001164817
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr12_+_62654120
|
0.432
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr6_-_109777085
|
0.432
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr6_-_46293215
|
0.426
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr22_-_43010894
|
0.423
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr19_+_50169080
|
0.420
|
|
BCL2L12
|
BCL2-like 12 (proline rich)
|
|
chr5_-_139682657
|
0.419
|
|
|
|
|
chr1_-_149783910
|
0.417
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr16_+_69726751
|
0.414
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr15_+_91478498
|
0.412
|
|
UNC45A
|
unc-45 homolog A (C. elegans)
|
|
chr13_+_97874540
|
0.406
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr8_-_109799769
|
0.405
|
NM_153015
|
TMEM74
|
transmembrane protein 74
|
|
chr18_-_47017940
|
0.405
|
NM_001199355
|
RPL17-C18ORF32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chr15_+_91478214
|
0.403
|
NM_018671
|
UNC45A
|
unc-45 homolog A (C. elegans)
|
|
chr16_-_74808699
|
0.399
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr1_-_68962726
|
0.390
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr11_-_31832758
|
0.390
|
|
PAX6
|
paired box 6
|
|
chr11_-_62369213
|
0.389
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chrM_+_5245
|
0.385
|
|
|
|
|
chr6_+_26217145
|
0.384
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr9_+_92219926
|
0.384
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr13_-_77460432
|
0.382
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|