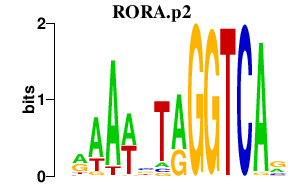
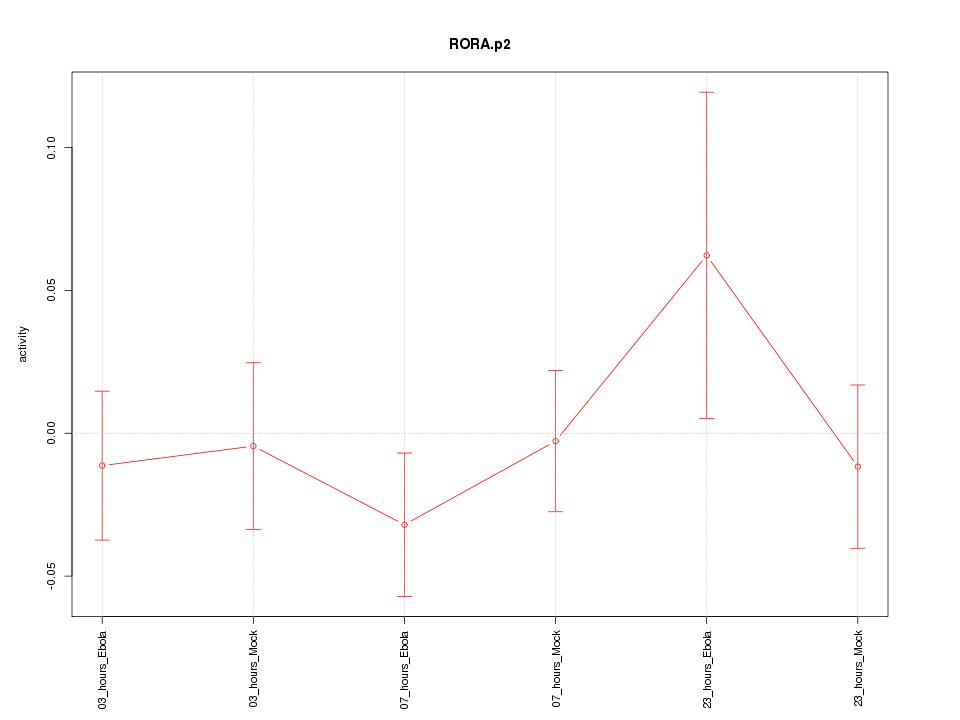
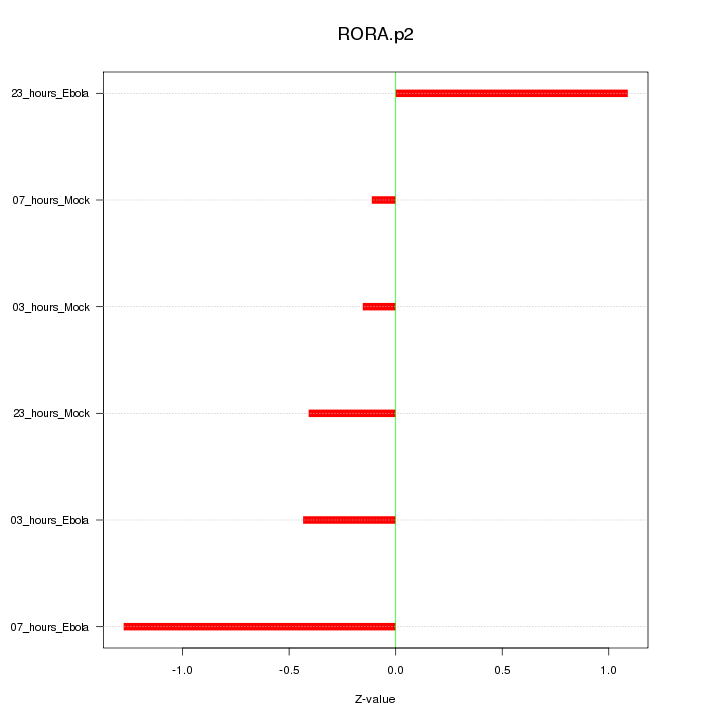
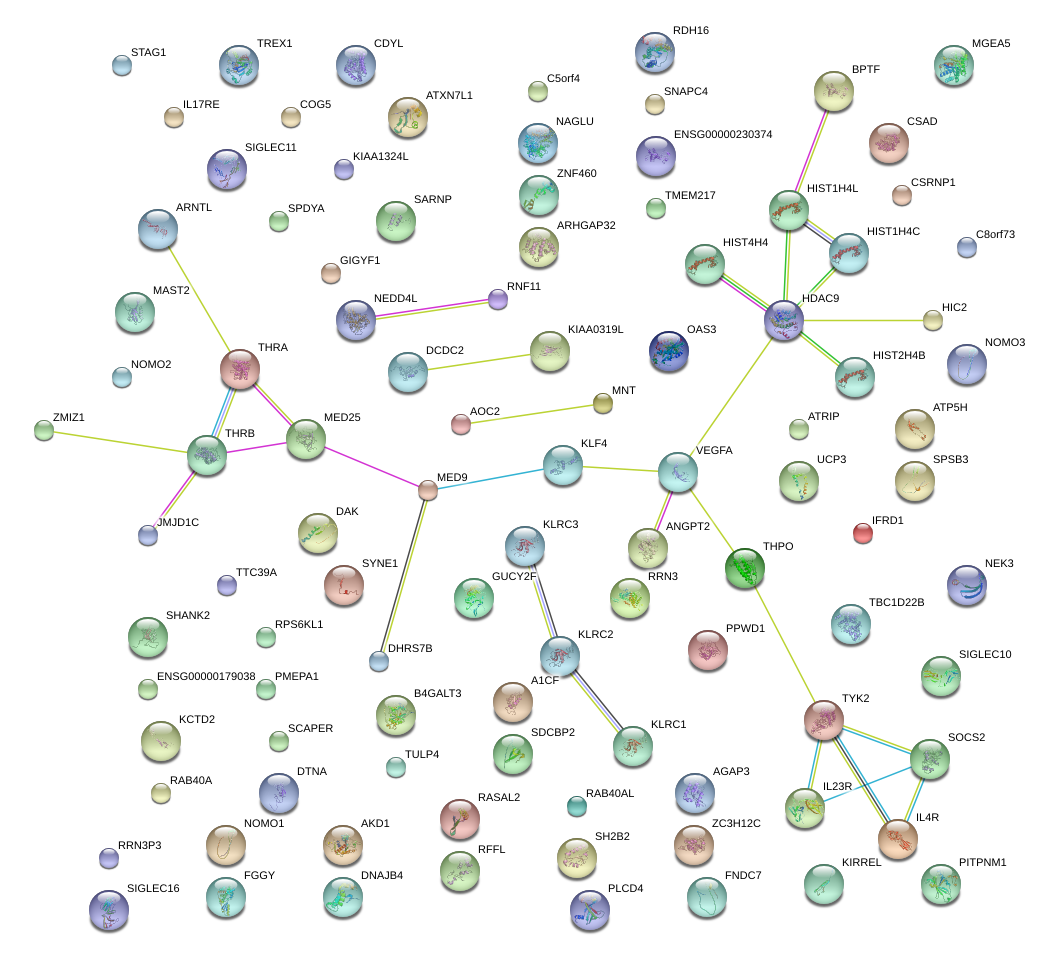
|
chr7_+_18535351
|
2.779
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr12_+_93963597
|
1.730
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr7_+_101928417
|
1.113
|
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr7_+_101928390
|
1.083
|
NM_020979
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr6_+_37252166
|
0.989
|
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr2_+_29038862
|
0.973
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr10_-_103578172
|
0.944
|
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase)
|
|
chr18_+_32073245
|
0.894
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr10_-_103578207
|
0.891
|
NM_001142434
NM_012215
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase)
|
|
chr10_-_103578179
|
0.881
|
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase)
|
|
chr10_-_103578181
|
0.877
|
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase)
|
|
chr1_+_46379258
|
0.838
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr18_+_32073343
|
0.829
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr16_-_1832576
|
0.818
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr16_-_1832304
|
0.776
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr17_+_40996596
|
0.613
|
NM_001158
NM_009590
|
AOC2
|
amine oxidase, copper containing 2 (retina-specific)
|
|
chr7_-_100286869
|
0.613
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr12_-_53575121
|
0.600
|
|
CSAD
|
cysteine sulfinic acid decarboxylase
|
|
chr16_-_1832571
|
0.584
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr7_-_107204705
|
0.570
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr11_+_13299260
|
0.541
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr18_+_55816546
|
0.539
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_+_74920329
|
0.532
|
|
|
|
|
chr1_+_67632168
|
0.511
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr11_-_70672861
|
0.478
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr6_+_43738757
|
0.475
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr11_+_13299349
|
0.472
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr11_-_67271839
|
0.455
|
|
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1
|
|
chr3_-_24536176
|
0.421
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr17_-_2304217
|
0.406
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr11_+_109963524
|
0.389
|
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr3_-_24536452
|
0.366
|
|
THRB
|
thyroid hormone receptor, beta
|
|
chr2_+_219472618
|
0.358
|
|
PLCD4
|
phospholipase C, delta 4
|
|
chr8_-_6420454
|
0.350
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr11_+_13299376
|
0.349
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr11_+_13299329
|
0.338
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr6_+_43738394
|
0.332
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chrX_-_102774416
|
0.326
|
NM_080879
|
RAB40A
|
RAB40A, member RAS oncogene family
|
|
chr3_-_39195034
|
0.320
|
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr1_+_109255555
|
0.306
|
NM_001144937
|
FNDC7
|
fibronectin type III domain containing 7
|
|
chr10_-_52645430
|
0.283
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr16_+_27325269
|
0.280
|
|
IL4R
|
interleukin 4 receptor
|
|
chr12_-_10588543
|
0.277
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr6_+_26104103
|
0.266
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chrX_-_114797017
|
0.265
|
|
|
|
|
chr16_-_15188089
|
0.261
|
NM_018427
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae)
|
|
chr9_-_110251308
|
0.260
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr8_-_144654921
|
0.253
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr11_+_13299391
|
0.241
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr16_-_21831730
|
0.227
|
|
RRN3P1
|
RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene 1
|
|
chr6_+_43737937
|
0.201
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr11_-_73720281
|
0.200
|
NM_003356
NM_022803
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr10_-_65028825
|
0.190
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr11_+_13299372
|
0.186
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chrX_-_108725284
|
0.182
|
NM_001522
|
GUCY2F
|
guanylate cyclase 2F, retinal
|
|
chr1_+_178310605
|
0.182
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr15_-_77176191
|
0.180
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr17_-_33390676
|
0.176
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr16_+_16326478
|
0.160
|
|
NOMO2
|
NODAL modulator 2
|
|
chr6_+_158734476
|
0.159
|
|
TULP4
|
tubby like protein 4
|
|
chr13_-_52733995
|
0.151
|
NM_001146099
NM_002498
|
NEK3
|
NIMA (never in mitosis gene a)-related kinase 3
|
|
chr1_+_157963062
|
0.146
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr11_-_129062092
|
0.145
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr7_-_105319608
|
0.145
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chrX_+_56755666
|
0.139
|
|
LOC550643
|
uncharacterized LOC550643
|
|
chr1_+_78470593
|
0.137
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr16_+_16326488
|
0.136
|
|
NOMO2
|
NODAL modulator 2
|
|
chr14_-_75389144
|
0.132
|
NM_031464
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1
|
|
chr12_-_56211503
|
0.126
|
|
SARNP
|
SAP domain containing ribonucleoprotein
|
|
chr16_+_16326471
|
0.125
|
|
NOMO2
|
NODAL modulator 2
|
|
chr1_-_35920081
|
0.124
|
|
KIAA0319L
|
KIAA0319-like
|
|
chr2_+_219472631
|
0.124
|
|
PLCD4
|
phospholipase C, delta 4
|
|
chr20_-_1309782
|
0.123
|
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr6_-_24383519
|
0.122
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr5_-_154230148
|
0.121
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr1_+_51701943
|
0.118
|
NM_014372
|
RNF11
|
ring finger protein 11
|
|
chr22_+_21771661
|
0.116
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr3_-_136471186
|
0.115
|
NM_005862
|
STAG1
|
stromal antigen 1
|
|
chr7_-_86595203
|
0.107
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr17_-_73043079
|
0.105
|
|
ATP5H
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d
|
|
chr6_+_43738177
|
0.104
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr19_-_10491172
|
0.104
|
|
TYK2
|
tyrosine kinase 2
|
|
chr17_+_65916163
|
0.103
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr12_-_57351381
|
0.103
|
NM_003708
|
RDH16
|
retinol dehydrogenase 16 (all-trans)
|
|
chr3_+_141105283
|
0.100
|
|
|
|
|
chr6_-_152623213
|
0.099
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr1_-_161147757
|
0.098
|
NM_001199873
NM_001199874
NM_003779
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr12_-_56211484
|
0.095
|
|
SARNP
|
SAP domain containing ribonucleoprotein
|
|
chr5_+_64859098
|
0.094
|
NM_015342
|
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1
|
|
chr9_-_41954687
|
0.091
|
|
MGC21881
|
uncharacterized locus MGC21881
|
|
chr3_+_48488113
|
0.091
|
|
ATRIP
|
ATR interacting protein
|
|
chr7_+_112063131
|
0.089
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr2_+_219472487
|
0.089
|
NM_032726
|
PLCD4
|
phospholipase C, delta 4
|
|
chr7_+_150782881
|
0.082
|
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr19_-_50464306
|
0.081
|
NM_001135163
NM_052884
|
SIGLEC11
|
sialic acid binding Ig-like lectin 11
|
|
chr6_-_37225894
|
0.077
|
NM_001162900
|
TMEM217
|
transmembrane protein 217
|
|
chr1_-_51810693
|
0.074
|
NM_001144832
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr17_+_40688264
|
0.071
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr16_+_1833003
|
0.071
|
|
|
|
|
chr12_+_113376301
|
0.069
|
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa
|
|
chr3_-_184095927
|
0.068
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr6_+_4836331
|
0.068
|
NM_001143970
|
CDYL
|
chromodomain protein, Y-like
|
|
chr16_+_16326454
|
0.067
|
|
NOMO2
|
NODAL modulator 2
|
|
chr3_+_9944295
|
0.066
|
NM_153483
|
IL17RE
|
interleukin 17 receptor E
|
|
chr19_+_57791794
|
0.064
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr3_+_48488180
|
0.063
|
NM_032166
NM_130384
|
TREX1
ATRIP
|
three prime repair exonuclease 1
ATR interacting protein
|
|
chr12_+_113376001
|
0.062
|
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa
|
|
chr9_-_139292888
|
0.061
|
NM_003086
|
SNAPC4
|
small nuclear RNA activating complex, polypeptide 4, 190kDa
|
|
chr19_+_50321503
|
0.059
|
NM_030973
|
MED25
|
mediator complex subunit 25
|
|
chr10_+_81070578
|
0.056
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr17_+_73043319
|
0.053
|
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr11_+_61102035
|
0.052
|
|
DAK
|
dihydroxyacetone kinase 2 homolog (S. cerevisiae)
|
|
chr1_+_59775749
|
0.051
|
NM_001244714
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr1_-_103496768
|
0.050
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr15_-_75135479
|
0.050
|
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr9_+_123884043
|
0.049
|
|
CNTRL
|
centriolin
|
|
chr12_+_65672682
|
0.048
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr6_-_134373618
|
0.048
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr15_-_75135480
|
0.048
|
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr17_+_40688189
|
0.045
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr6_-_30181197
|
0.045
|
NM_001242783
NM_003449
|
TRIM26
|
tripartite motif containing 26
|
|
chr5_+_137688270
|
0.044
|
NM_016604
|
KDM3B
|
lysine (K)-specific demethylase 3B
|
|
chr19_+_55996537
|
0.042
|
NM_020378
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative)
|
|
chr15_-_66084438
|
0.039
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr15_+_45422130
|
0.039
|
NM_017434
NM_175940
|
DUOX1
|
dual oxidase 1
|
|
chr1_-_205744543
|
0.038
|
NM_001135662
NM_001135663
NM_001135664
NM_003929
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1
|
|
chr10_+_18041217
|
0.037
|
NM_001098844
|
TMEM236
|
transmembrane protein 236
|
|
chr1_-_19217384
|
0.036
|
NM_001161504
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr22_+_21738039
|
0.035
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr15_+_102501826
|
0.033
|
|
LOC100134445
WASH1
WASH3P
|
uncharacterized LOC100134445
WAS protein family homolog 1
WAS protein family homolog 3 pseudogene
|
|
chr12_+_65672422
|
0.030
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr10_-_105845619
|
0.030
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr6_+_143929316
|
0.029
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr10_+_114043413
|
0.029
|
NM_058222
|
TECTB
|
tectorin beta
|
|
chr9_-_139342563
|
0.028
|
|
SEC16A
|
SEC16 homolog A (S. cerevisiae)
|
|
chr22_-_20427814
|
0.028
|
|
PI4KAP1
|
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 1
|
|
chr7_+_115863004
|
0.026
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr11_-_72380107
|
0.024
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr16_-_12009918
|
0.024
|
|
GSPT1
|
G1 to S phase transition 1
|
|
chr17_-_73043040
|
0.024
|
NM_001003785
NM_006356
|
ATP5H
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d
|
|
chr3_+_23987514
|
0.023
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chrX_-_23925947
|
0.021
|
NM_024122
|
APOO
|
apolipoprotein O
|
|
chr17_-_26697358
|
0.021
|
NM_000638
|
VTN
|
vitronectin
|
|
chrM_+_5769
|
0.021
|
|
|
|
|
chr7_+_152161119
|
0.020
|
|
LOC100128822
|
uncharacterized LOC100128822
|
|
chr2_+_27851514
|
0.019
|
NM_001145047
NM_001145048
NM_001145049
|
GPN1
|
GPN-loop GTPase 1
|
|
chr17_+_26989193
|
0.019
|
|
SUPT6H
|
suppressor of Ty 6 homolog (S. cerevisiae)
|
|
chr10_-_52645384
|
0.019
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr12_+_105153258
|
0.019
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr1_+_31883020
|
0.018
|
NM_001199037
|
SERINC2
|
serine incorporator 2
|
|
chr15_-_75135429
|
0.018
|
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr1_+_28764660
|
0.016
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr10_-_105845556
|
0.015
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr5_+_66124603
|
0.015
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr1_+_51701914
|
0.013
|
|
RNF11
|
ring finger protein 11
|
|
chr2_-_216003068
|
0.012
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr6_+_31913920
|
0.010
|
|
CFB
|
complement factor B
|
|
chr11_-_47615902
|
0.010
|
NM_031909
|
C1QTNF4
|
C1q and tumor necrosis factor related protein 4
|
|
chr1_+_171750807
|
0.010
|
|
METTL13
|
methyltransferase like 13
|
|
chr6_-_111927082
|
0.009
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr6_-_35696359
|
0.009
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr3_+_132318980
|
0.008
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chrX_-_132092224
|
0.007
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr4_+_52917440
|
0.007
|
NM_145263
|
SPATA18
|
spermatogenesis associated 18 homolog (rat)
|
|
chr11_-_18258383
|
0.006
|
NM_006512
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr12_+_60083125
|
0.005
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr5_+_61708565
|
0.001
|
NM_016338
|
IPO11
|
importin 11
|
|
chr7_-_42971779
|
0.000
|
NM_002787
|
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2
|