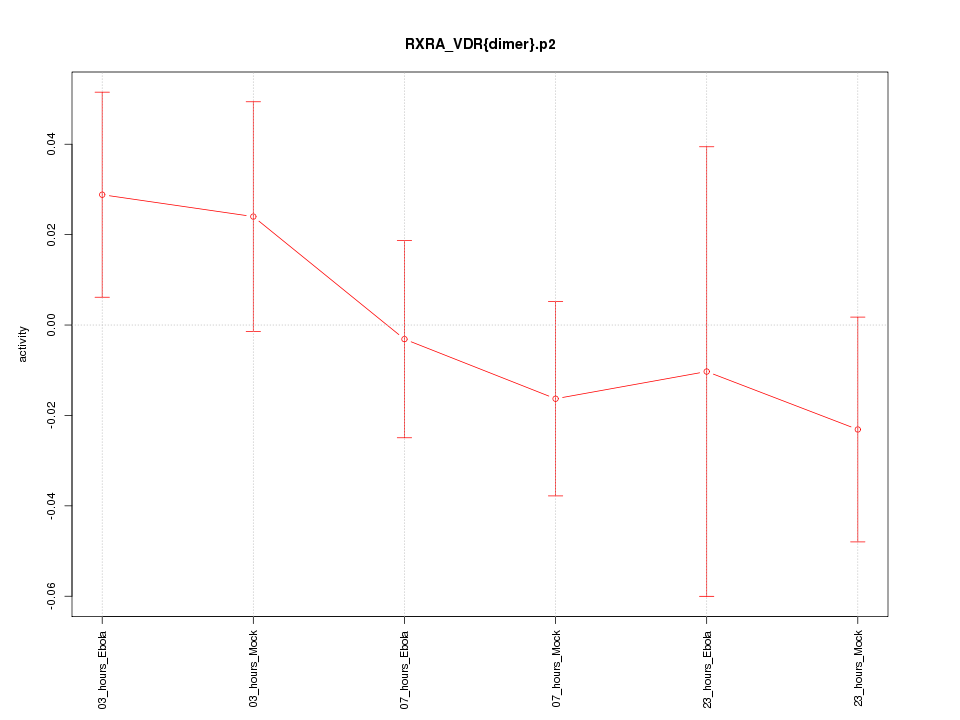
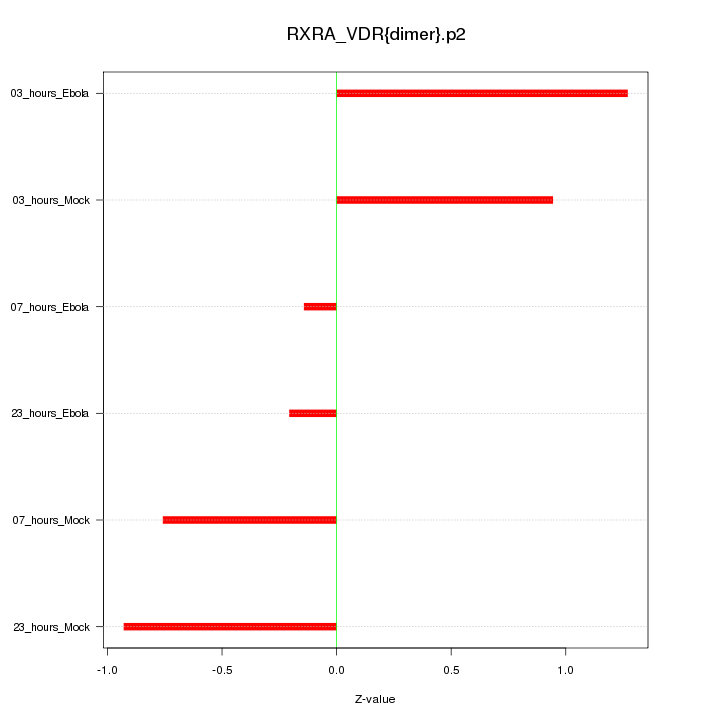
|
chr1_+_100111430
|
0.523
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr11_+_124609841
|
0.404
|
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr3_-_27764197
|
0.398
|
|
EOMES
|
eomesodermin
|
|
chr16_+_19079220
|
0.396
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr7_+_143701021
|
0.384
|
NM_001005281
|
OR6B1
|
olfactory receptor, family 6, subfamily B, member 1
|
|
chr16_+_19078916
|
0.355
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr3_-_151987332
|
0.330
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr1_+_226736603
|
0.322
|
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr20_+_42193754
|
0.317
|
NM_001199264
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr16_+_19078958
|
0.317
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr19_+_42817434
|
0.293
|
NM_173633
|
TMEM145
|
transmembrane protein 145
|
|
chr8_+_99956630
|
0.270
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr20_+_3801164
|
0.252
|
NM_001204446
NM_001204447
NM_018347
|
C20orf29
|
chromosome 20 open reading frame 29
|
|
chr2_+_230787195
|
0.248
|
NM_174899
|
FBXO36
|
F-box protein 36
|
|
chr2_-_25194772
|
0.241
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr9_+_131644780
|
0.235
|
NM_001127244
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr11_+_73087670
|
0.229
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr19_+_7985193
|
0.222
|
NM_003083
|
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa
|
|
chr2_+_25264913
|
0.220
|
NM_014971
|
EFR3B
|
EFR3 homolog B (S. cerevisiae)
|
|
chr19_+_589849
|
0.217
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr1_+_61869699
|
0.209
|
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_226736500
|
0.208
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr18_-_59560303
|
0.204
|
NM_173557
|
RNF152
|
ring finger protein 152
|
|
chr5_+_99871234
|
0.203
|
|
FAM174A
|
family with sequence similarity 174, member A
|
|
chr10_-_33623978
|
0.199
|
|
NRP1
|
neuropilin 1
|
|
chr19_-_14260728
|
0.198
|
|
LPHN1
|
latrophilin 1
|
|
chr1_+_200993041
|
0.194
|
|
|
|
|
chr2_-_25194484
|
0.188
|
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr10_-_33623771
|
0.182
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr12_-_72667194
|
0.173
|
|
LOC283392
|
uncharacterized LOC283392
|
|
chr17_+_7554711
|
0.157
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr19_+_7985884
|
0.143
|
|
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa
|
|
chr6_-_42016383
|
0.142
|
|
CCND3
|
cyclin D3
|
|
chr6_+_35227478
|
0.137
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr7_-_122339186
|
0.135
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr18_+_48086402
|
0.133
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr12_-_77459198
|
0.133
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr2_+_238768186
|
0.132
|
NM_005855
|
RAMP1
|
receptor (G protein-coupled) activity modifying protein 1
|
|
chr20_-_39946119
|
0.132
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr19_+_13250072
|
0.131
|
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr2_+_233498206
|
0.129
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chrX_-_49121171
|
0.129
|
NM_001114377
NM_014009
|
FOXP3
|
forkhead box P3
|
|
chr17_-_79791152
|
0.128
|
NM_001093767
NM_207368
|
FAM195B
|
family with sequence similarity 195, member B
|
|
chr19_-_58858900
|
0.127
|
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr15_-_89764809
|
0.125
|
NM_000326
|
RLBP1
|
retinaldehyde binding protein 1
|
|
chr14_+_55168831
|
0.124
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr14_+_105780751
|
0.124
|
NM_001100913
NM_015197
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr3_-_71114006
|
0.123
|
NM_001244813
|
FOXP1
|
forkhead box P1
|
|
chr6_-_42016609
|
0.121
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr12_-_95397487
|
0.120
|
NM_018838
|
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12
|
|
chr3_-_125655881
|
0.119
|
NM_001015050
NM_001195223
|
ALG1L
|
asparagine-linked glycosylation 1-like
|
|
chr3_-_27763784
|
0.118
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr9_-_77643024
|
0.118
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr13_+_51796363
|
0.116
|
NM_001242312
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr14_+_95047730
|
0.114
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr6_-_132272311
|
0.114
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr19_-_12595547
|
0.112
|
|
ZNF709
|
zinc finger protein 709
|
|
chr1_-_153585504
|
0.112
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr21_+_40177754
|
0.111
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr4_+_144435076
|
0.111
|
|
|
|
|
chr3_+_38032260
|
0.110
|
|
VILL
|
villin-like
|
|
chr4_+_134073180
|
0.109
|
|
PCDH10
|
protocadherin 10
|
|
chr3_-_127309313
|
0.108
|
|
TPRA1
|
transmembrane protein, adipocyte asscociated 1
|
|
chr12_-_109251333
|
0.105
|
NM_001161330
NM_018984
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr1_-_203317366
|
0.104
|
|
FMOD
|
fibromodulin
|
|
chr1_-_33366838
|
0.103
|
NM_033504
|
TMEM54
|
transmembrane protein 54
|
|
chr10_+_111767877
|
0.101
|
|
ADD3
|
adducin 3 (gamma)
|
|
chr3_-_14988939
|
0.100
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr14_-_101351183
|
0.100
|
NM_001134888
|
RTL1
|
retrotransposon-like 1
|
|
chr1_+_151043079
|
0.099
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr3_-_14988976
|
0.096
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr9_+_34458810
|
0.094
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr21_+_40177847
|
0.094
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr13_+_24844846
|
0.093
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr2_+_74740585
|
0.092
|
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr1_+_44398991
|
0.092
|
NM_001136215
NM_057090
NM_057091
|
ARTN
|
artemin
|
|
chr9_-_77643156
|
0.092
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr16_+_77756388
|
0.092
|
NM_001105663
NM_001243657
NM_001243660
NM_001243661
|
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7
|
|
chrX_+_48336379
|
0.091
|
NM_177434
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr3_-_59035757
|
0.090
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr1_+_228353814
|
0.090
|
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae)
|
|
chr1_-_3816823
|
0.090
|
NM_207356
|
C1orf174
|
chromosome 1 open reading frame 174
|
|
chr17_-_33905556
|
0.089
|
NM_000286
|
PEX12
|
peroxisomal biogenesis factor 12
|
|
chr15_-_55881049
|
0.088
|
NM_015617
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr1_-_3816704
|
0.088
|
|
C1orf174
|
chromosome 1 open reading frame 174
|
|
chr8_-_23540401
|
0.087
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr1_-_33338081
|
0.086
|
NM_001171941
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr1_-_55331350
|
0.085
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chrX_+_106161589
|
0.083
|
NM_001171095
|
CLDN2
|
claudin 2
|
|
chrX_+_46940235
|
0.082
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr14_-_93581507
|
0.082
|
|
ITPK1
|
inositol-tetrakisphosphate 1-kinase
|
|
chr9_+_116638544
|
0.081
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr6_+_39760138
|
0.079
|
|
|
|
|
chr11_+_64126616
|
0.079
|
NM_001006944
NM_003942
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4
|
|
chr14_+_94640648
|
0.078
|
NM_020958
NM_058237
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4
|
|
chr4_+_144434555
|
0.078
|
NM_003601
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr20_+_42875943
|
0.078
|
|
GDAP1L1
|
ganglioside-induced differentiation-associated protein 1-like 1
|
|
chr15_-_40633122
|
0.077
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr14_+_24439082
|
0.076
|
NM_001193636
NM_001193637
NM_001193635
|
DHRS4L2
DHRS4L1
|
dehydrogenase/reductase (SDR family) member 4 like 2
dehydrogenase/reductase (SDR family) member 4 like 1
|
|
chr11_-_67188614
|
0.075
|
|
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme
|
|
chr1_-_1850704
|
0.074
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr7_+_143078358
|
0.073
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr20_-_43024303
|
0.071
|
|
|
|
|
chr17_+_48046561
|
0.070
|
NM_138281
|
DLX4
|
distal-less homeobox 4
|
|
chr16_+_57653909
|
0.070
|
NM_001145770
NM_005682
|
GPR56
|
G protein-coupled receptor 56
|
|
chr7_+_143078457
|
0.070
|
|
ZYX
|
zyxin
|
|
chr1_-_33336289
|
0.068
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr7_+_44795786
|
0.066
|
NM_174929
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr12_-_111126894
|
0.066
|
NM_032369
|
HVCN1
|
hydrogen voltage-gated channel 1
|
|
chr3_+_156392369
|
0.066
|
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr6_-_32145841
|
0.065
|
NM_032741
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr17_-_76778306
|
0.065
|
NM_004762
NM_017456
|
CYTH1
|
cytohesin 1
|
|
chr3_-_59035737
|
0.064
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr3_+_184032414
|
0.064
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr18_+_21452981
|
0.063
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr11_+_7535213
|
0.063
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr7_+_143078444
|
0.063
|
|
ZYX
|
zyxin
|
|
chr3_+_184032243
|
0.063
|
NM_001194946
NM_001194947
NM_198241
NM_198242
NM_198244
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_95397455
|
0.063
|
|
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12
|
|
chr10_+_119806334
|
0.063
|
|
CASC2
|
cancer susceptibility candidate 2 (non-protein coding)
|
|
chr17_+_65373362
|
0.063
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr22_+_45705766
|
0.062
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr2_-_200320732
|
0.061
|
|
SATB2
|
SATB homeobox 2
|
|
chr19_-_47987410
|
0.061
|
|
KPTN
|
kaptin (actin binding protein)
|
|
chr10_-_30316338
|
0.061
|
|
KIAA1462
|
KIAA1462
|
|
chr10_+_102732285
|
0.060
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr17_+_75447325
|
0.059
|
|
SEPT9
|
septin 9
|
|
chr2_+_113341999
|
0.059
|
NM_032309
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr3_+_184032849
|
0.058
|
NM_182917
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr16_-_67184822
|
0.058
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr6_+_18155535
|
0.058
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr10_+_30723050
|
0.058
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr22_+_45705965
|
0.058
|
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr3_+_51741080
|
0.057
|
NM_000839
NM_001130063
|
GRM2
|
glutamate receptor, metabotropic 2
|
|
chr2_-_204399906
|
0.057
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr6_+_34204928
|
0.056
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr22_+_39077960
|
0.056
|
|
TOMM22
|
translocase of outer mitochondrial membrane 22 homolog (yeast)
|
|
chr12_+_52306112
|
0.055
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr11_+_101918168
|
0.055
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr2_+_128458943
|
0.055
|
|
SFT2D3
|
SFT2 domain containing 3
|
|
chr9_+_131644427
|
0.054
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr1_+_161284165
|
0.053
|
NM_001035511
NM_001035512
NM_001035513
NM_003001
|
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa
|
|
chr22_+_39077953
|
0.053
|
NM_020243
|
TOMM22
|
translocase of outer mitochondrial membrane 22 homolog (yeast)
|
|
chr4_-_74904329
|
0.053
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr19_+_38042275
|
0.053
|
NM_152606
|
LOC100507433
ZNF540
|
uncharacterized LOC100507433
zinc finger protein 540
|
|
chr19_-_42759151
|
0.052
|
|
ERF
|
Ets2 repressor factor
|
|
chr19_+_10654591
|
0.052
|
NM_032885
|
ATG4D
|
ATG4 autophagy related 4 homolog D (S. cerevisiae)
|
|
chr13_+_93879011
|
0.052
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr5_-_7869113
|
0.052
|
NM_024091
|
FASTKD3
|
FAST kinase domains 3
|
|
chr1_-_41487386
|
0.052
|
NM_001168247
NM_144990
|
SLFNL1
|
schlafen-like 1
|
|
chr2_+_66666445
|
0.050
|
|
MEIS1
|
Meis homeobox 1
|
|
chr3_+_184032319
|
0.050
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr22_+_39077975
|
0.050
|
|
TOMM22
|
translocase of outer mitochondrial membrane 22 homolog (yeast)
|
|
chr1_+_203764802
|
0.050
|
|
ZC3H11A
|
zinc finger CCCH-type containing 11A
|
|
chr1_-_221915426
|
0.049
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr21_+_45937499
|
0.049
|
|
C21orf90
|
chromosome 21 open reading frame 90
|
|
chr10_-_21785873
|
0.049
|
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr3_+_156392204
|
0.048
|
NM_015508
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr9_+_35673852
|
0.048
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr1_-_221915393
|
0.048
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr4_-_25032306
|
0.048
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr15_+_50474357
|
0.048
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr19_-_12595625
|
0.047
|
NM_152601
|
ZNF709
|
zinc finger protein 709
|
|
chr10_+_104503724
|
0.047
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr20_+_42194736
|
0.047
|
NM_016276
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr11_-_28129687
|
0.046
|
|
KIF18A
|
kinesin family member 18A
|
|
chrX_+_49019180
|
0.046
|
NM_001099681
NM_001099682
NM_024859
|
MAGIX
|
MAGI family member, X-linked
|
|
chr10_+_80899458
|
0.045
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr17_-_6616651
|
0.045
|
NM_001143838
NM_177550
|
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5
|
|
chr6_+_39760148
|
0.045
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr7_+_91763851
|
0.044
|
|
LOC613126
|
uncharacterized LOC613126
|
|
chr9_+_128024097
|
0.044
|
|
GAPVD1
|
GTPase activating protein and VPS9 domains 1
|
|
chr11_-_77791264
|
0.043
|
NM_001203260
NM_001203261
NM_001203262
NM_001204054
NM_001204055
NM_004549
|
NDUFC2-KCTD14
NDUFC2
|
NDUFC2-KCTD14 readthrough
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa
|
|
chr21_-_35986744
|
0.043
|
NM_203417
|
RCAN1
|
regulator of calcineurin 1
|
|
chr16_-_3767449
|
0.043
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr9_+_128024070
|
0.043
|
|
GAPVD1
|
GTPase activating protein and VPS9 domains 1
|
|
chr9_+_131644368
|
0.042
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr20_-_60982334
|
0.042
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr6_-_167040700
|
0.042
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr19_+_45394642
|
0.041
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr10_+_104503631
|
0.041
|
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_+_32859509
|
0.041
|
NM_001039111
|
TRIM71
|
tripartite motif containing 71
|
|
chr7_-_148787866
|
0.041
|
NM_152411
|
ZNF786
|
zinc finger protein 786
|
|
chr9_+_128024109
|
0.040
|
NM_015635
|
GAPVD1
|
GTPase activating protein and VPS9 domains 1
|
|
chr10_-_13390018
|
0.040
|
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr1_+_145663156
|
0.040
|
|
RNF115
|
ring finger protein 115
|
|
chr13_+_50760015
|
0.040
|
|
|
|
|
chr19_-_42759279
|
0.039
|
NM_006494
|
ERF
|
Ets2 repressor factor
|
|
chr12_-_109251271
|
0.039
|
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr11_-_28129646
|
0.038
|
|
KIF18A
|
kinesin family member 18A
|
|
chr11_-_28129701
|
0.037
|
NM_031217
|
KIF18A
|
kinesin family member 18A
|
|
chr14_-_23445885
|
0.037
|
|
AJUBA
|
ajuba LIM protein
|
|
chr3_+_29322802
|
0.037
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr2_+_70485204
|
0.037
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chr3_+_57541973
|
0.037
|
NM_177966
|
PDE12
|
phosphodiesterase 12
|
|
chrX_-_21676416
|
0.036
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr18_-_74844710
|
0.036
|
|
MBP
|
myelin basic protein
|
|
chr19_-_17559375
|
0.036
|
NM_001190844
|
TMEM221
|
transmembrane protein 221
|
|
chr7_+_97841565
|
0.036
|
NM_177455
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr17_+_79935410
|
0.035
|
NM_001251888
NM_024083
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|