|
chr13_-_110438896
|
3.872
|
NM_003749
|
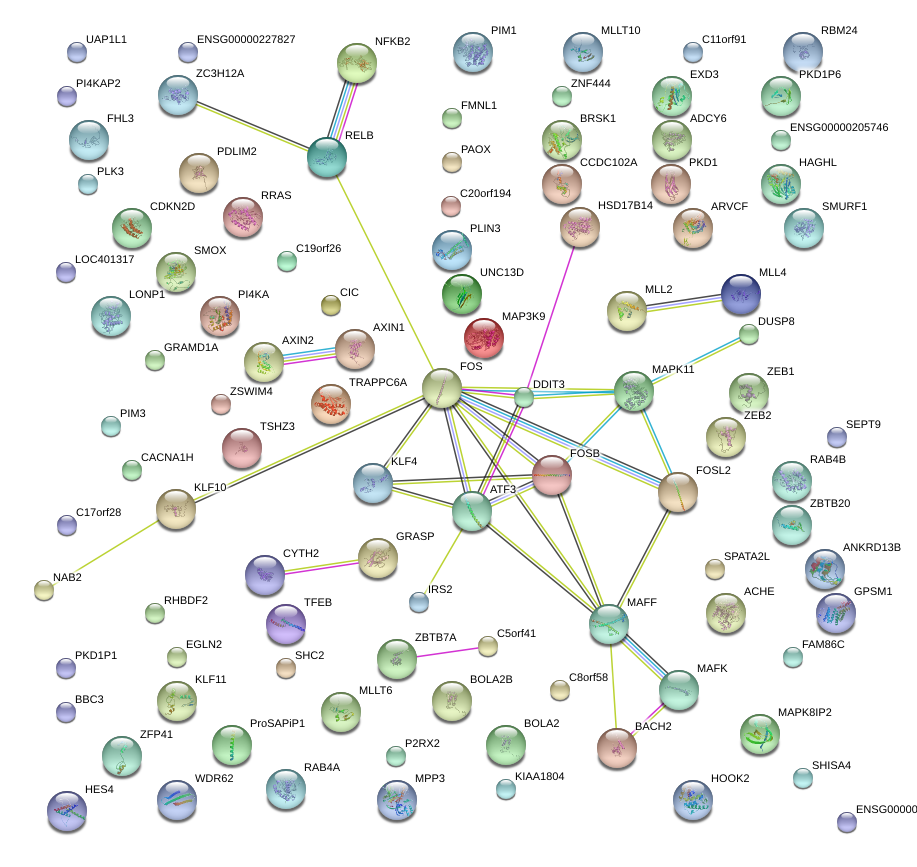
IRS2
|
insulin receptor substrate 2
|
|
chr1_+_212781969
|
3.148
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr19_-_10679328
|
3.037
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr10_+_135192694
|
3.001
|
NM_152911
NM_207127
NM_207128
|
PAOX
|
polyamine oxidase (exo-N4-amino)
|
|
chr19_-_47734215
|
2.954
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr9_+_139972195
|
2.776
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr1_-_935469
|
2.751
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr9_-_110251754
|
2.724
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr21_-_37528578
|
2.723
|
|
LOC100506428
|
uncharacterized LOC100506428
|
|
chr1_-_38471170
|
2.710
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr10_+_104155431
|
2.654
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr1_+_212782103
|
2.561
|
|
ATF3
|
activating transcription factor 3
|
|
chr8_+_22457098
|
2.457
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr22_-_50708720
|
2.369
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr6_+_37137882
|
2.349
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr9_-_140317583
|
2.321
|
|
EXD3
|
exonuclease 3'-5' domain containing 3
|
|
chr19_-_49339667
|
2.276
|
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr19_+_42788605
|
2.271
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr14_+_75745480
|
2.269
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr16_+_1203240
|
2.266
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr16_-_402617
|
2.261
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr19_+_41305100
|
2.175
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_+_45504686
|
2.161
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr6_-_91006460
|
2.132
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr9_+_139221931
|
2.072
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr17_+_43299277
|
2.068
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr12_-_57914287
|
2.035
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr19_-_10679620
|
2.010
|
NM_001800
NM_079421
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr12_+_57482886
|
2.009
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr19_+_41305333
|
1.986
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_-_45681454
|
1.978
|
NM_024108
|
TRAPPC6A
|
trafficking protein particle complex 6A
|
|
chr19_+_41305041
|
1.969
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_-_12886263
|
1.927
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr6_-_41702734
|
1.920
|
NM_007162
|
TFEB
|
transcription factor EB
|
|
chr5_+_172484353
|
1.917
|
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr19_-_4065467
|
1.915
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr19_+_55795766
|
1.912
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr1_+_201857997
|
1.910
|
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr20_-_3388219
|
1.901
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr7_+_28448970
|
1.880
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr19_-_50143350
|
1.869
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr1_+_201857789
|
1.867
|
NM_198149
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr20_-_3154169
|
1.856
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr7_-_100493481
|
1.852
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr17_-_74497406
|
1.841
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr2_+_28616360
|
1.835
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_+_1570321
|
1.827
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr17_-_72968790
|
1.821
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr6_+_17281773
|
1.818
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr9_+_139971938
|
1.812
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr1_+_37940129
|
1.791
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr1_+_37940180
|
1.788
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr16_-_89768094
|
1.776
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr3_-_114866090
|
1.768
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_+_144328990
|
1.764
|
|
ZFP41
|
zinc finger protein 41 homolog (mouse)
|
|
chr16_-_2185824
|
1.762
|
NM_000296
NM_001009944
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr1_+_37940074
|
1.758
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr17_-_72968750
|
1.752
|
|
|
|
|
chr2_+_10183630
|
1.745
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr19_+_7660698
|
1.742
|
NM_001080429
NM_020902
|
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3
|
|
chr1_-_1475739
|
1.723
|
NM_001114748
|
TMEM240
|
transmembrane protein 240
|
|
chr22_+_51039178
|
1.712
|
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr16_+_1203737
|
1.708
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr16_-_57570455
|
1.698
|
NM_033212
|
CCDC102A
|
coiled-coil domain containing 102A
|
|
chr9_-_35115844
|
1.697
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr19_-_31840118
|
1.694
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr22_-_20004251
|
1.691
|
NM_001670
|
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome
|
|
chr1_-_935352
|
1.672
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr19_+_45971249
|
1.655
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr2_-_145274916
|
1.637
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr16_+_776957
|
1.634
|
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr19_-_1237840
|
1.627
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr19_+_41284161
|
1.624
|
|
RAB4B-EGLN2
RAB4B
|
RAB4B-EGLN2 readthrough
RAB4B, member RAS oncogene family
|
|
chr1_+_45266048
|
1.619
|
|
PLK3
|
polo-like kinase 3
|
|
chr19_+_36208917
|
1.614
|
NM_014727
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr8_-_103666018
|
1.610
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr16_-_402398
|
1.609
|
|
AXIN1
|
axin 1
|
|
chr12_-_49182718
|
1.605
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr1_+_45265896
|
1.603
|
|
PLK3
|
polo-like kinase 3
|
|
chr2_+_28615695
|
1.584
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr11_-_1593149
|
1.563
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr19_+_13906207
|
1.556
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr19_-_49339913
|
1.547
|
NM_016246
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr14_-_71275732
|
1.535
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr19_+_36209125
|
1.523
|
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr2_+_149633173
|
1.512
|
|
|
|
|
chr16_-_29606639
|
1.508
|
|
|
|
|
chr12_+_133195365
|
1.500
|
NM_012226
NM_016318
NM_170682
NM_170683
NM_174872
NM_174873
|
P2RX2
|
purinergic receptor P2X, ligand-gated ion channel, 2
|
|
chr1_+_233463513
|
1.498
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chr19_+_48972574
|
1.495
|
|
CYTH2
|
cytohesin 2
|
|
chr22_-_20427814
|
1.493
|
|
PI4KAP1
|
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 1
|
|
chr17_-_63557615
|
1.488
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr22_+_50354428
|
1.481
|
|
PIM3
|
pim-3 oncogene
|
|
chr11_-_33722285
|
1.480
|
NM_001166692
|
C11orf91
|
chromosome 11 open reading frame 91
|
|
chr20_-_3388586
|
1.472
|
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr16_-_29466284
|
1.470
|
NM_001031827
NM_001039182
|
BOLA2
BOLA2B
|
bolA homolog 2 (E. coli)
bolA homolog 2B (E. coli)
|
|
chr17_+_27920479
|
1.461
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr12_+_52400725
|
1.459
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr22_+_51039100
|
1.451
|
NM_012324
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr22_+_38597938
|
1.448
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr20_+_4129409
|
1.443
|
NM_175839
NM_175840
NM_175841
NM_175842
|
SMOX
|
spermine oxidase
|
|
chr2_+_28615771
|
1.442
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr19_+_56652529
|
1.440
|
NM_001253792
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr8_+_8086103
|
1.439
|
|
FLJ10661
|
family with sequence similarity 86, member A pseudogene
|
|
chr19_+_35491329
|
1.433
|
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr12_+_57482674
|
1.427
|
NM_005967
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr19_-_4867705
|
1.424
|
NM_001164189
NM_001164194
NM_005817
|
PLIN3
|
perilipin 3
|
|
chr17_-_41910439
|
1.423
|
NM_001932
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr2_+_28616346
|
1.412
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr17_+_36861398
|
1.406
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr16_-_29606286
|
1.399
|
|
LOC440354
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) pseudogene
|
|
chr2_+_28615668
|
1.399
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr6_-_91006580
|
1.397
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr7_-_98741375
|
1.382
|
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr19_-_460995
|
1.380
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr3_-_39195034
|
1.379
|
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr19_+_35491225
|
1.377
|
NM_001136199
NM_020895
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr1_+_45266035
|
1.377
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr3_+_184097860
|
1.368
|
NM_003741
|
CHRD
|
chordin
|
|
chr22_+_31477231
|
1.362
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr4_-_967269
|
1.361
|
|
DGKQ
|
diacylglycerol kinase, theta 110kDa
|
|
chr10_-_31320772
|
1.356
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr3_-_39195101
|
1.355
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr11_+_64073040
|
1.355
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr17_+_7255207
|
1.355
|
NM_001002914
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr19_+_56652689
|
1.337
|
|
ZNF444
|
zinc finger protein 444
|
|
chr19_+_4969123
|
1.332
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr8_+_144329197
|
1.329
|
|
ZFP41
|
zinc finger protein 41 homolog (mouse)
|
|
chr17_+_79989485
|
1.326
|
NM_005052
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3)
|
|
chr8_+_81398416
|
1.320
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr19_+_2164119
|
1.319
|
NM_032482
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr7_+_64498643
|
1.319
|
|
CCT6P3
|
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 3
|
|
chr22_+_38598065
|
1.317
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr19_+_55795533
|
1.317
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr20_-_62711160
|
1.315
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr22_-_41032684
|
1.313
|
NM_020831
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr19_+_47759802
|
1.312
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr16_-_122573
|
1.311
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr6_-_43596919
|
1.305
|
NM_019096
|
GTPBP2
|
GTP binding protein 2
|
|
chr6_-_33285493
|
1.304
|
NM_005453
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr10_-_126849066
|
1.303
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr19_-_4867638
|
1.303
|
|
PLIN3
|
perilipin 3
|
|
chr2_+_42275017
|
1.300
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr11_-_46142955
|
1.297
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr17_-_43394326
|
1.297
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr19_-_4867655
|
1.295
|
|
PLIN3
|
perilipin 3
|
|
chr17_+_26698666
|
1.295
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr15_+_41136622
|
1.293
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr1_-_16302272
|
1.291
|
NM_001242884
NM_003443
|
ZBTB17
|
zinc finger and BTB domain containing 17
|
|
chr8_-_103667882
|
1.286
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr22_-_41032623
|
1.285
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr20_-_31172874
|
1.285
|
|
LOC284804
|
uncharacterized LOC284804
|
|
chr17_-_63557764
|
1.279
|
|
AXIN2
|
axin 2
|
|
chrX_+_153665258
|
1.274
|
NM_001493
|
GDI1
|
GDP dissociation inhibitor 1
|
|
chr19_-_344778
|
1.270
|
NM_017550
|
MIER2
|
mesoderm induction early response 1, family member 2
|
|
chr6_-_33285718
|
1.268
|
NM_001145338
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr17_-_80231240
|
1.260
|
|
CSNK1D
|
casein kinase 1, delta
|
|
chr7_-_86974693
|
1.256
|
|
TP53TG1
|
TP53 target 1 (non-protein coding)
|
|
chr5_+_172483285
|
1.255
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr17_-_42277319
|
1.255
|
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr8_+_144329105
|
1.255
|
NM_173832
|
ZFP41
|
zinc finger protein 41 homolog (mouse)
|
|
chr9_-_110252045
|
1.254
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr16_+_66638632
|
1.254
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr19_+_49375677
|
1.248
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr5_+_179234002
|
1.238
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr9_+_130159383
|
1.234
|
NM_014580
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8
|
|
chr6_+_33387846
|
1.234
|
NM_006772
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr19_+_49375679
|
1.233
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr9_-_35111534
|
1.229
|
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr19_+_36266366
|
1.220
|
NM_001172630
NM_052948
|
ARHGAP33
|
Rho GTPase activating protein 33
|
|
chr6_+_2245965
|
1.217
|
|
|
|
|
chr12_+_132379262
|
1.214
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr1_+_68151092
|
1.213
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr19_-_1174263
|
1.212
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr11_-_67141008
|
1.202
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr11_-_61684981
|
1.201
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr6_+_43738757
|
1.201
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr19_-_46000159
|
1.200
|
NM_005619
NM_206900
|
RTN2
|
reticulon 2
|
|
chr7_+_18126543
|
1.197
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr7_+_150076405
|
1.197
|
NM_173680
|
ZNF775
|
zinc finger protein 775
|
|
chr19_+_49375638
|
1.196
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr16_+_89787951
|
1.196
|
NM_001113525
|
ZNF276
|
zinc finger protein 276
|
|
chr19_+_48673936
|
1.191
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr12_-_26277807
|
1.190
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr16_+_67562704
|
1.186
|
NM_024519
NM_001193522
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr1_+_28585959
|
1.185
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr4_-_82393025
|
1.184
|
NM_152545
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr22_+_50946627
|
1.183
|
NM_001185011
NM_014551
NM_152299
|
NCAPH2
|
non-SMC condensin II complex, subunit H2
|
|
chr19_-_18632857
|
1.181
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr2_-_27718059
|
1.181
|
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr19_-_51893791
|
1.181
|
NM_001193623
|
LOC147646
|
uncharacterized LOC147646
|
|
chr4_-_967327
|
1.181
|
NM_001347
|
DGKQ
|
diacylglycerol kinase, theta 110kDa
|
|
chr5_+_177540465
|
1.173
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr16_+_2039945
|
1.173
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr2_+_30369749
|
1.168
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr16_+_66638652
|
1.167
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr12_-_112819755
|
1.166
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr19_+_4969126
|
1.160
|
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr7_-_45128492
|
1.158
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr16_-_75285383
|
1.158
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|