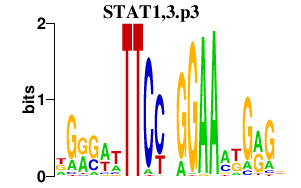
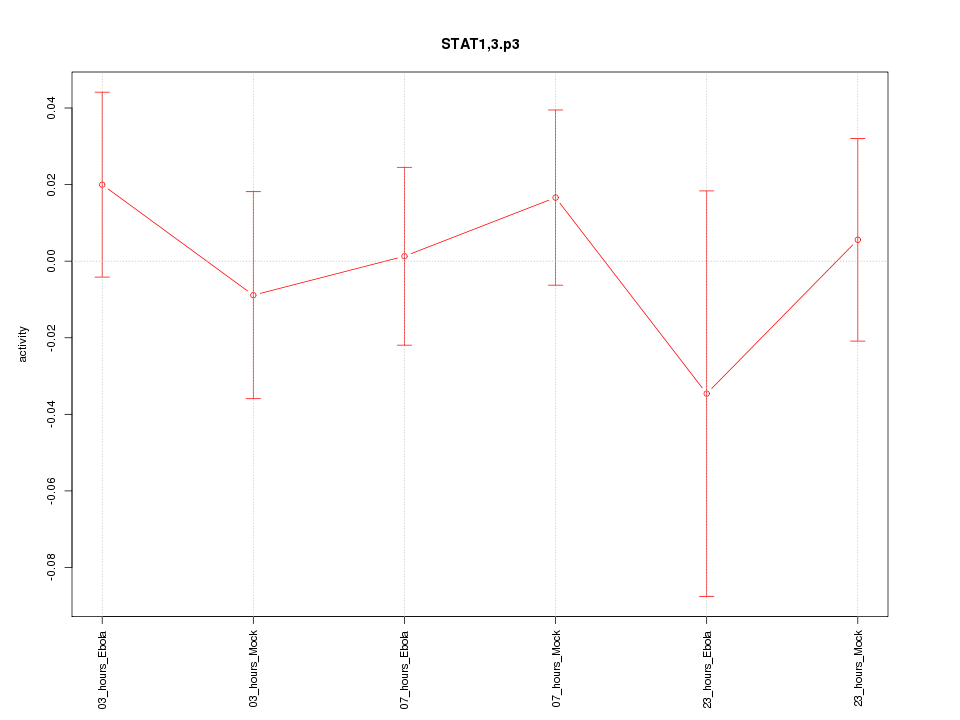
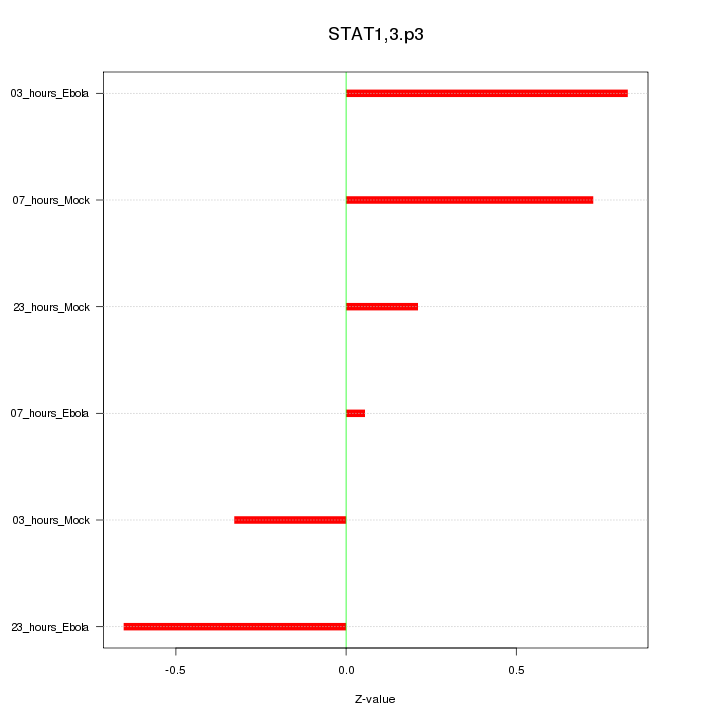
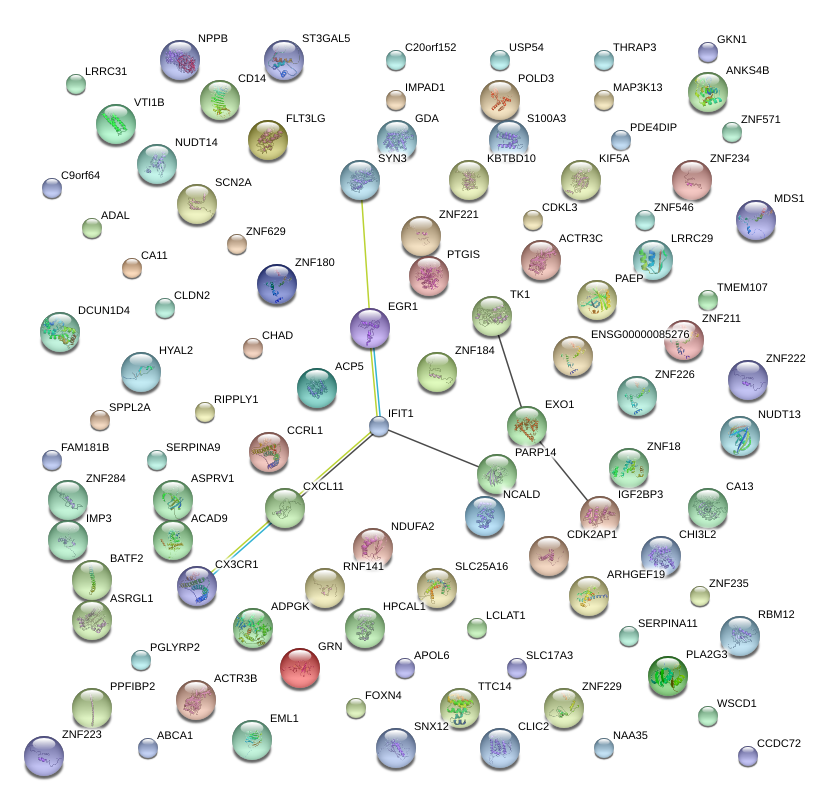
|
chr9_-_107666046
|
1.051
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_-_16539103
|
0.848
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr1_-_144994943
|
0.778
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_144994839
|
0.687
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_144994908
|
0.673
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr4_-_76957139
|
0.641
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr19_+_44645728
|
0.621
|
|
ZNF234
|
zinc finger protein 234
|
|
chr22_+_36044412
|
0.605
|
NM_030641
|
APOL6
|
apolipoprotein L, 6
|
|
chr8_-_102803196
|
0.561
|
|
NCALD
|
neurocalcin delta
|
|
chr14_+_100239935
|
0.548
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr19_+_44617510
|
0.490
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr17_-_8079607
|
0.437
|
NM_032354
NM_183065
|
TMEM107
|
transmembrane protein 107
|
|
chr3_-_50360109
|
0.414
|
NM_003773
NM_033158
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr19_+_44645708
|
0.412
|
NM_001144824
NM_006630
|
ZNF234
|
zinc finger protein 234
|
|
chr9_-_107665959
|
0.388
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr19_-_15590203
|
0.384
|
NM_052890
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr20_+_34556528
|
0.382
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr22_+_36044475
|
0.380
|
|
APOL6
|
apolipoprotein L, 6
|
|
chr19_+_44507076
|
0.377
|
NM_006300
|
ZNF230
|
zinc finger protein 230
|
|
chr19_+_40502942
|
0.348
|
NM_178544
|
ZNF546
|
zinc finger protein 546
|
|
chrX_-_106146546
|
0.342
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr19_+_44529493
|
0.330
|
NM_001129996
NM_013360
|
ZNF284
ZNF222
|
zinc finger protein 284
zinc finger protein 222
|
|
chr19_-_44952630
|
0.297
|
NM_014518
|
ZNF229
|
zinc finger protein 229
|
|
chr19_+_44576266
|
0.296
|
NM_001037813
|
ZNF284
|
zinc finger protein 284
|
|
chr16_-_30798206
|
0.273
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr11_+_7618416
|
0.270
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr8_+_86157732
|
0.270
|
|
CA13
|
carbonic anhydrase XIII
|
|
chr3_+_128598459
|
0.264
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr17_-_11900688
|
0.260
|
NM_144680
|
ZNF18
|
zinc finger protein 18
|
|
chr3_+_128598332
|
0.248
|
NM_014049
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr3_+_128598471
|
0.239
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr19_+_44556163
|
0.235
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr10_+_91152321
|
0.235
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr16_-_67260900
|
0.232
|
NM_001004055
|
LRRC29
|
leucine rich repeat containing 29
|
|
chr5_-_133706717
|
0.228
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr16_-_67260720
|
0.228
|
NM_012163
|
LRRC29
|
leucine rich repeat containing 29
|
|
chr19_-_44617271
|
0.224
|
|
LOC100379224
|
uncharacterized LOC100379224
|
|
chr15_-_75932509
|
0.222
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr3_+_128598440
|
0.221
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr3_+_128598520
|
0.219
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr3_+_128598497
|
0.210
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr2_-_70189396
|
0.203
|
NM_152792
|
ASPRV1
|
aspartic peptidase, retroviral-like 1
|
|
chr3_+_122399665
|
0.195
|
NM_017554
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr1_+_54519259
|
0.192
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr8_-_102803178
|
0.189
|
|
NCALD
|
neurocalcin delta
|
|
chrX_-_154563735
|
0.188
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr8_+_86158022
|
0.185
|
|
|
|
|
chr22_-_31536339
|
0.184
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr17_-_76183074
|
0.181
|
NM_003258
|
TK1
|
thymidine kinase 1, soluble
|
|
chr3_+_180319898
|
0.180
|
NM_001042601
NM_133462
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr1_-_153521717
|
0.178
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr19_+_44455379
|
0.174
|
NM_013359
|
ZNF221
|
zinc finger protein 221
|
|
chr12_-_123756164
|
0.171
|
NM_004642
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1
|
|
chr15_+_43622871
|
0.169
|
NM_001012969
NM_001159280
|
ADAL
|
adenosine deaminase-like
|
|
chr8_-_57906143
|
0.165
|
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr14_+_73957670
|
0.162
|
|
C14orf169
|
chromosome 14 open reading frame 169
|
|
chr19_-_44809126
|
0.160
|
NM_004234
|
ZNF235
|
zinc finger protein 235
|
|
chr1_-_11918889
|
0.160
|
NM_002521
|
NPPB
|
natriuretic peptide B
|
|
chr6_-_27440459
|
0.157
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr9_-_86571533
|
0.148
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr1_+_111770280
|
0.147
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr6_-_25874439
|
0.143
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr11_+_74303626
|
0.142
|
NM_006591
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit
|
|
chr22_-_33402648
|
0.140
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr3_+_48481706
|
0.139
|
|
CCDC72
|
coiled-coil domain containing 72
|
|
chr1_+_242012032
|
0.133
|
NM_003686
NM_006027
|
EXO1
|
exonuclease 1
|
|
chr8_-_57906423
|
0.131
|
NM_017813
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr15_-_73076113
|
0.131
|
NM_031284
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr11_+_62104882
|
0.130
|
|
ASRGL1
|
asparaginase like 1
|
|
chr20_-_48184652
|
0.122
|
NM_000961
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase
|
|
chr2_+_170366211
|
0.122
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr2_+_30670082
|
0.118
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr19_-_49149252
|
0.117
|
NM_001217
|
CA11
|
carbonic anhydrase XI
|
|
chr19_-_45004482
|
0.117
|
NM_013256
|
ZNF180
|
zinc finger protein 180
|
|
chr3_+_185000714
|
0.116
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr9_+_88556056
|
0.115
|
NM_024635
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr12_-_109747024
|
0.115
|
NM_213596
|
FOXN4
|
forkhead box N4
|
|
chr2_+_166150340
|
0.111
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr1_+_242011471
|
0.111
|
NM_130398
|
EXO1
|
exonuclease 1
|
|
chr14_-_94919085
|
0.110
|
NM_001080451
|
SERPINA11
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11
|
|
chr3_-_129118241
|
0.107
|
|
RPL32P3
|
ribosomal protein L32 pseudogene 3
|
|
chr11_+_62104773
|
0.107
|
NM_001083926
NM_025080
|
ASRGL1
|
asparaginase like 1
|
|
chr7_-_150000068
|
0.105
|
NM_001164459
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast)
|
|
chr8_-_57906402
|
0.104
|
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr5_-_140013285
|
0.104
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr9_-_86571631
|
0.104
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr10_+_74870209
|
0.102
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr9_+_74764382
|
0.102
|
|
GDA
|
guanine deaminase
|
|
chr14_-_105647520
|
0.101
|
NM_177533
|
NUDT14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14
|
|
chr4_+_52709165
|
0.101
|
NM_001040402
NM_015115
|
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr19_-_11689591
|
0.100
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr20_-_34243118
|
0.100
|
|
RBM12
|
RNA binding motif protein 12
|
|
chr3_+_132316080
|
0.099
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr19_+_58144510
|
0.099
|
NM_006385
NM_198855
|
ZNF211
|
zinc finger protein 211
|
|
chr3_-_168865521
|
0.097
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr17_+_5972425
|
0.096
|
|
WSCD1
|
WSC domain containing 1
|
|
chr1_+_36690038
|
0.096
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr2_-_86094792
|
0.096
|
NM_001042437
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chrX_+_106143393
|
0.095
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr14_-_68141328
|
0.094
|
NM_006370
|
VTI1B
|
vesicle transport through interaction with t-SNAREs homolog 1B (yeast)
|
|
chr17_+_42422490
|
0.093
|
NM_002087
|
GRN
|
granulin
|
|
chr15_-_73076071
|
0.088
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr15_-_73076062
|
0.088
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr19_+_49977465
|
0.087
|
NM_001204502
NM_001459
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr17_-_48546174
|
0.086
|
NM_001267
|
CHAD
|
chondroadherin
|
|
chr3_-_169587598
|
0.086
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr5_+_139944040
|
0.085
|
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr14_-_92414004
|
0.084
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr12_+_56211862
|
0.078
|
|
ORMDL2
|
ORM1-like 2 (S. cerevisiae)
|
|
chr11_+_111944967
|
0.078
|
NM_001082969
NM_001082970
NM_018195
|
C11orf57
|
chromosome 11 open reading frame 57
|
|
chr10_-_38146978
|
0.077
|
|
ZNF248
|
zinc finger protein 248
|
|
chr1_-_212004113
|
0.077
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr9_+_88556102
|
0.076
|
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr1_-_166845469
|
0.075
|
|
TADA1
|
transcriptional adaptor 1
|
|
chr3_+_58223228
|
0.075
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr10_-_61469322
|
0.075
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr11_+_48002259
|
0.074
|
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr15_-_73076025
|
0.073
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr15_-_73076057
|
0.073
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr4_+_119809978
|
0.072
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr8_-_57906376
|
0.067
|
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr3_-_48481518
|
0.066
|
NM_024661
|
CCDC51
|
coiled-coil domain containing 51
|
|
chrX_-_100548044
|
0.064
|
NM_024885
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa
|
|
chr15_-_64385988
|
0.064
|
NM_001014812
NM_032231
|
FAM96A
|
family with sequence similarity 96, member A
|
|
chr8_+_25316714
|
0.064
|
|
CDCA2
|
cell division cycle associated 2
|
|
chr9_+_74764394
|
0.064
|
|
GDA
|
guanine deaminase
|
|
chr12_-_69357019
|
0.063
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr6_+_74405505
|
0.060
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr9_+_5450455
|
0.060
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr19_-_59084089
|
0.060
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr2_+_17935379
|
0.059
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr12_+_56211718
|
0.058
|
NM_014182
|
ORMDL2
|
ORM1-like 2 (S. cerevisiae)
|
|
chr1_+_207262583
|
0.058
|
NM_000716
NM_001017364
NM_001017367
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr6_+_54711568
|
0.057
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr5_-_179498995
|
0.057
|
NM_018434
|
RNF130
|
ring finger protein 130
|
|
chr17_+_76183397
|
0.057
|
NM_001010982
NM_001145526
|
AFMID
|
arylformamidase
|
|
chr15_+_90808833
|
0.057
|
NM_001033088
|
NGRN
|
neugrin, neurite outgrowth associated
|
|
chr6_+_139349818
|
0.056
|
NM_021243
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr8_-_18666295
|
0.054
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr12_-_122231554
|
0.054
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chrX_+_48432740
|
0.053
|
NM_006743
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3
|
|
chr1_-_160617042
|
0.052
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr16_-_56553960
|
0.051
|
NM_031885
|
BBS2
|
Bardet-Biedl syndrome 2
|
|
chr16_-_67517715
|
0.051
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr11_-_118966175
|
0.051
|
NM_002105
|
H2AFX
|
H2A histone family, member X
|
|
chr1_-_202927269
|
0.050
|
|
ADIPOR1
|
adiponectin receptor 1
|
|
chr8_+_22853360
|
0.049
|
NM_001160037
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr1_-_202927410
|
0.049
|
|
ADIPOR1
|
adiponectin receptor 1
|
|
chrX_+_119738542
|
0.049
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr8_+_25316736
|
0.048
|
|
CDCA2
|
cell division cycle associated 2
|
|
chr20_+_61436160
|
0.047
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr14_-_76127158
|
0.046
|
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr8_+_96037186
|
0.046
|
NM_152416
|
C8orf38
|
chromosome 8 open reading frame 38
|
|
chr17_+_71229357
|
0.045
|
|
C17orf80
|
chromosome 17 open reading frame 80
|
|
chr11_+_75479766
|
0.044
|
NM_032564
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr7_-_135662034
|
0.044
|
|
MTPN
|
myotrophin
|
|
chr19_-_5839702
|
0.042
|
NM_000150
NM_001040701
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr5_+_109025085
|
0.042
|
|
|
|
|
chr12_+_57916607
|
0.042
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr19_+_35168558
|
0.041
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr1_+_233086319
|
0.041
|
NM_032324
|
NTPCR
|
nucleoside-triphosphatase, cancer-related
|
|
chr12_-_75603214
|
0.040
|
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr6_-_28304151
|
0.040
|
NM_001243241
NM_001243242
NM_001243244
NM_030899
|
ZNF323
|
zinc finger protein 323
|
|
chr16_-_4897265
|
0.039
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr5_-_139944045
|
0.039
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr12_-_7261788
|
0.039
|
NM_016546
|
C1RL
|
complement component 1, r subcomponent-like
|
|
chr12_-_122839015
|
0.039
|
|
CLIP1
|
CAP-GLY domain containing linker protein 1
|
|
chr1_-_163113938
|
0.039
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr11_+_75479802
|
0.039
|
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr7_-_151511925
|
0.039
|
NM_001040633
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr1_-_226497415
|
0.038
|
|
LIN9
|
lin-9 homolog (C. elegans)
|
|
chr5_+_134209426
|
0.037
|
NM_024715
|
TXNDC15
|
thioredoxin domain containing 15
|
|
chr13_-_99738639
|
0.037
|
NM_001130049
NM_015296
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr5_-_140012730
|
0.035
|
|
CD14
|
CD14 molecule
|
|
chr15_-_42386657
|
0.034
|
NM_178034
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic)
|
|
chr16_-_1470709
|
0.034
|
|
C16orf91
|
chromosome 16 open reading frame 91
|
|
chr10_+_72575724
|
0.033
|
|
SGPL1
|
sphingosine-1-phosphate lyase 1
|
|
chr6_-_28303910
|
0.033
|
NM_001135216
NM_001243243
|
ZNF323
|
zinc finger protein 323
|
|
chr1_+_45965855
|
0.033
|
NM_015506
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria
|
|
chr1_+_61869699
|
0.032
|
|
NFIA
|
nuclear factor I/A
|
|
chr12_+_102271082
|
0.032
|
NM_018370
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr22_+_25003610
|
0.032
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr11_-_61197455
|
0.032
|
NM_001136040
NM_001142565
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa
|
|
chrX_-_134185978
|
0.031
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chr12_+_104324186
|
0.030
|
NM_003299
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr3_-_118959723
|
0.029
|
NM_003778
NM_212543
|
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4
|
|
chr20_+_37101457
|
0.029
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr6_+_53883713
|
0.029
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr19_+_17416476
|
0.028
|
NM_023937
|
MRPL34
|
mitochondrial ribosomal protein L34
|
|
chr17_-_74349591
|
0.027
|
|
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1
|
|
chr5_+_73109342
|
0.027
|
NM_001244364
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr19_+_49977817
|
0.027
|
NM_001204503
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr1_+_233086372
|
0.026
|
|
NTPCR
|
nucleoside-triphosphatase, cancer-related
|
|
chr16_+_4545858
|
0.026
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr13_-_95953572
|
0.026
|
NM_001105515
NM_005845
|
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4
|
|
chr6_+_150690027
|
0.026
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr5_+_140261792
|
0.026
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr1_+_156123321
|
0.024
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr5_-_139943829
|
0.024
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr9_-_75567915
|
0.024
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|