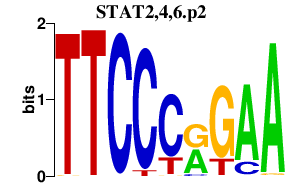
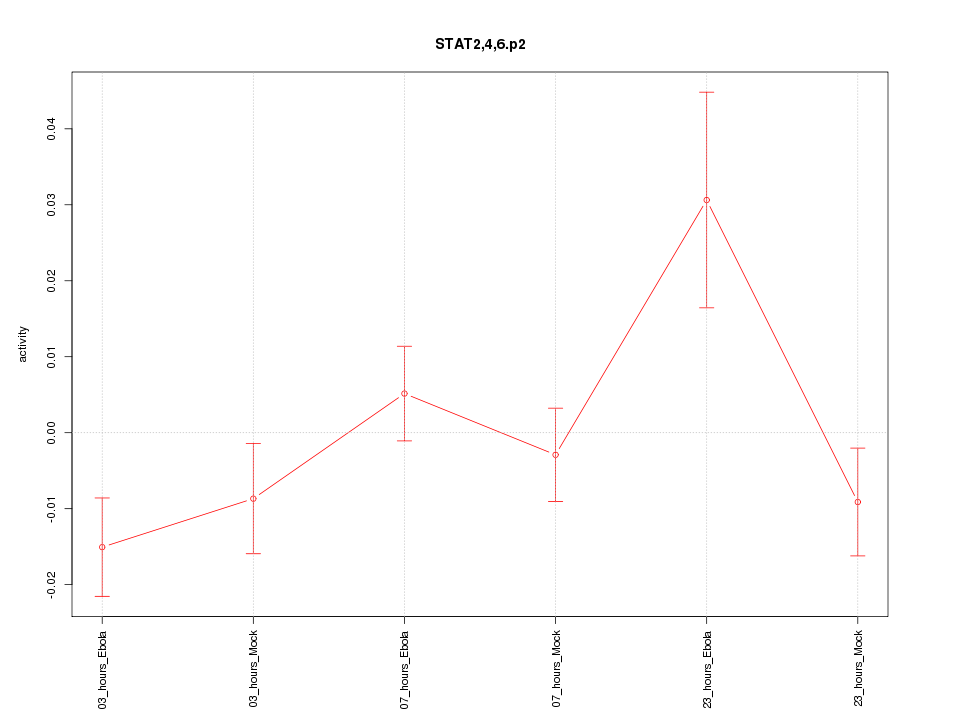
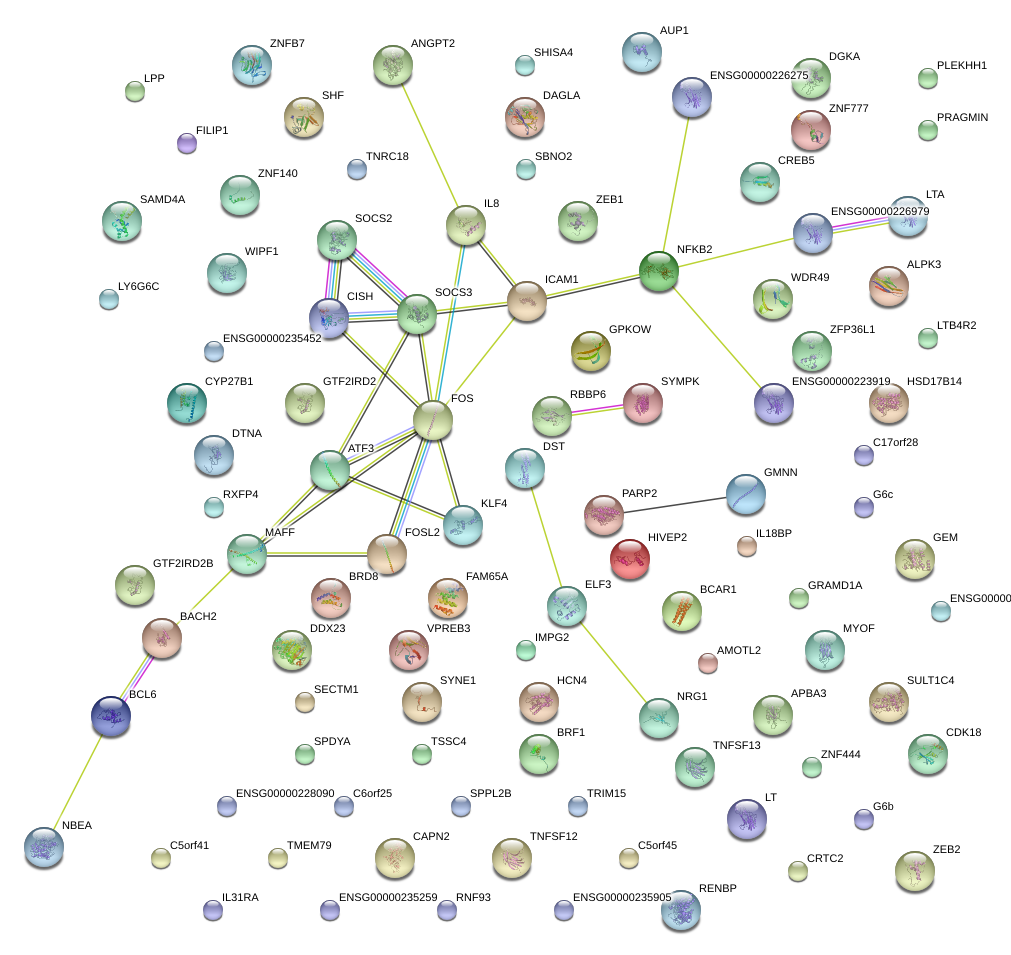
|
chr19_-_49339667
|
1.795
|
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr8_+_32405727
|
1.774
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr17_-_76356153
|
1.695
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr5_+_172483285
|
1.657
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr6_-_56716713
|
1.614
|
|
DST
|
dystonin
|
|
chr2_-_145277879
|
1.576
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr14_+_75745480
|
1.572
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr22_+_38598065
|
1.415
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr3_-_187463227
|
1.396
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr8_-_8239188
|
1.388
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr2_-_145277568
|
1.370
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr22_+_38597938
|
1.260
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr15_+_85359910
|
1.196
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr19_-_49339913
|
1.144
|
NM_016246
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr3_-_187463474
|
1.123
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr8_-_95274540
|
1.107
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr16_-_3109335
|
1.099
|
|
LOC100507419
|
uncharacterized LOC100507419
|
|
chr6_-_56716412
|
1.046
|
|
DST
|
dystonin
|
|
chr7_+_28530747
|
1.043
|
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_-_91006460
|
1.035
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr4_+_74606222
|
1.026
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr11_+_71710971
|
1.013
|
NM_001145055
|
IL18BP
|
interleukin 18 binding protein
|
|
chr7_-_149157866
|
1.011
|
NM_015694
|
ZNF777
|
zinc finger protein 777
|
|
chr9_-_110251308
|
1.007
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr11_+_71710662
|
1.002
|
NM_001145057
|
IL18BP
|
interleukin 18 binding protein
|
|
chr8_-_95274520
|
0.992
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr1_+_201979689
|
0.961
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr14_+_24779356
|
0.959
|
NM_001164692
NM_019839
|
LTB4R2
|
leukotriene B4 receptor 2
|
|
chr10_+_104154228
|
0.914
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr12_-_58159386
|
0.904
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr1_+_212788359
|
0.895
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr3_-_101039403
|
0.894
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr19_-_1132109
|
0.819
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr8_+_32579350
|
0.818
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr9_-_110251754
|
0.814
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr12_+_93964801
|
0.812
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr19_+_10381763
|
0.807
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr1_-_153930911
|
0.804
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr2_+_108994366
|
0.800
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr1_+_156254069
|
0.797
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr14_+_55168831
|
0.796
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr8_-_6420454
|
0.790
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr13_+_36050885
|
0.763
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr19_+_10381516
|
0.763
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr19_-_13947101
|
0.759
|
|
LOC284454
|
uncharacterized LOC284454
|
|
chr10_-_95241943
|
0.758
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chrX_-_153210057
|
0.753
|
NM_002910
|
RENBP
|
renin binding protein
|
|
chr2_+_29038862
|
0.751
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr5_-_137514299
|
0.751
|
NM_001164326
NM_006696
NM_139199
|
BRD8
|
bromodomain containing 8
|
|
chr6_+_31540092
|
0.749
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr16_-_75285383
|
0.734
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr17_-_72968790
|
0.733
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr6_+_30130982
|
0.724
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr1_+_201979747
|
0.721
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr6_-_76203256
|
0.720
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr3_-_167371701
|
0.716
|
|
WDR49
|
WD repeat domain 49
|
|
chr6_-_31689455
|
0.704
|
NM_025261
|
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr1_+_155911479
|
0.704
|
NM_181885
|
RXFP4
|
relaxin/insulin-like family peptide receptor 4
|
|
chr1_-_153931077
|
0.704
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr16_+_24567628
|
0.700
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr17_+_7462064
|
0.692
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr2_-_74756817
|
0.683
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr3_-_134093235
|
0.677
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr11_+_61447816
|
0.675
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chrX_-_48980070
|
0.672
|
NM_015698
|
GPKOW
|
G patch domain and KOW motifs
|
|
chr6_-_91006580
|
0.669
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr19_+_56652689
|
0.669
|
|
ZNF444
|
zinc finger protein 444
|
|
chr1_+_223900328
|
0.667
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr11_+_2421983
|
0.663
|
|
TSSC4
|
tumor suppressing subtransferable candidate 4
|
|
chr7_-_104654443
|
0.660
|
|
LOC100216545
|
uncharacterized LOC100216545
|
|
chr17_-_72968750
|
0.660
|
|
|
|
|
chr14_+_20811712
|
0.653
|
NM_001042618
NM_005484
|
PARP2
|
poly (ADP-ribose) polymerase 2
|
|
chr1_+_201857789
|
0.649
|
NM_198149
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr2_+_28616360
|
0.642
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr1_-_153931096
|
0.642
|
NM_181715
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr15_-_73661604
|
0.641
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chrX_-_73834427
|
0.640
|
|
|
|
|
chr3_+_187870808
|
0.639
|
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr16_+_67571342
|
0.633
|
NM_001193524
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr5_+_55149149
|
0.628
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr14_+_67999915
|
0.625
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr19_-_46366337
|
0.617
|
NM_004819
|
SYMPK
|
symplekin
|
|
chr1_+_205473768
|
0.617
|
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr17_+_47923271
|
0.614
|
NM_001242791
|
FLJ45513
|
uncharacterized LOC729220
|
|
chr19_+_35491329
|
0.614
|
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr15_-_45493372
|
0.612
|
NM_138356
|
SHF
|
Src homology 2 domain containing F
|
|
chr22_-_24096551
|
0.609
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr19_-_3761621
|
0.605
|
NM_004886
|
APBA3
|
amyloid beta (A4) precursor protein-binding, family A, member 3
|
|
chr12_+_56324945
|
0.604
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr19_+_35491225
|
0.604
|
NM_001136199
NM_020895
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr6_-_152702450
|
0.602
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr14_-_105715740
|
0.602
|
NM_001242789
|
BRF1
|
BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae)
|
|
chr1_+_201857997
|
0.598
|
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr19_+_2328679
|
0.594
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr5_-_179285791
|
0.593
|
NM_001017987
NM_016175
|
C5orf45
|
chromosome 5 open reading frame 45
|
|
chr18_+_32173281
|
0.587
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr3_-_50649110
|
0.587
|
NM_013324
NM_145071
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr1_+_223900212
|
0.585
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr12_+_133656984
|
0.585
|
NM_003440
|
ZNF140
|
zinc finger protein 140
|
|
chr7_-_5463176
|
0.583
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr19_+_56652529
|
0.583
|
NM_001253792
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr12_-_49245904
|
0.576
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr7_+_102004286
|
0.571
|
NM_001242713
|
LOC100289561
LOC100630923
|
uncharacterized LOC100289561
LOC100289561-PRKRIP1 readthrough
|
|
chr7_+_74508346
|
0.567
|
NM_001003795
|
GTF2IRD2B
GTF2IRD2
|
GTF2I repeat domain containing 2B
GTF2I repeat domain containing 2
|
|
chr17_-_80291610
|
0.566
|
|
SECTM1
|
secreted and transmembrane 1
|
|
chr6_-_30654406
|
0.563
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr2_-_175499275
|
0.562
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr6_+_31691120
|
0.562
|
NM_025260
NM_138272
NM_138273
NM_138274
NM_138275
NM_138277
|
C6orf25
|
chromosome 6 open reading frame 25
|
|
chr9_-_130186491
|
0.560
|
|
|
|
|
chr1_+_223900226
|
0.560
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr6_-_143266283
|
0.559
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr10_+_104155431
|
0.552
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr22_+_38599026
|
0.550
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr9_+_92219926
|
0.547
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr19_+_48111437
|
0.547
|
NM_015711
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1
|
|
chr9_+_102584136
|
0.544
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr12_-_93964636
|
0.539
|
|
|
|
|
chr6_-_43595046
|
0.538
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr2_-_208994408
|
0.536
|
NM_020989
|
CRYGC
|
crystallin, gamma C
|
|
chr16_-_3085541
|
0.534
|
NM_001103175
|
CCDC64B
|
coiled-coil domain containing 64B
|
|
chr6_-_33756885
|
0.533
|
|
LEMD2
|
LEM domain containing 2
|
|
chr1_+_160336856
|
0.533
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr21_+_30452872
|
0.532
|
NM_020152
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr8_-_124665165
|
0.525
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr19_+_2476122
|
0.524
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr6_-_30654978
|
0.524
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chrX_-_73834460
|
0.520
|
NM_016120
NM_183353
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr14_-_57960432
|
0.518
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr16_-_74455239
|
0.517
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr11_-_65667809
|
0.510
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_+_2328628
|
0.510
|
NM_001077238
NM_152988
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr3_+_122296432
|
0.509
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr14_+_65453947
|
0.507
|
|
|
|
|
chr1_+_223900061
|
0.507
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr6_-_111136407
|
0.506
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr12_-_39299335
|
0.506
|
NM_153634
|
CPNE8
|
copine VIII
|
|
chr20_+_51589752
|
0.505
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr22_-_31503484
|
0.505
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr1_+_203274646
|
0.503
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr11_-_125550674
|
0.501
|
NM_001612
NM_020069
NM_020107
NM_020108
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chr21_+_35736322
|
0.499
|
NM_172201
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2
|
|
chr19_-_40724272
|
0.496
|
NM_152479
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr19_+_11200124
|
0.494
|
|
LDLR
|
low density lipoprotein receptor
|
|
chr7_-_37025696
|
0.492
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr19_+_2328685
|
0.490
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr6_+_144471631
|
0.490
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr1_+_151138497
|
0.488
|
NM_001204856
NM_024041
|
SCNM1
|
sodium channel modifier 1
|
|
chr2_+_74757127
|
0.485
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr22_+_29138018
|
0.485
|
NM_172002
|
HSCB
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr9_-_4152182
|
0.485
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr11_-_89956121
|
0.485
|
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr1_+_110453232
|
0.483
|
NM_000757
NM_172210
NM_172212
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr8_+_144680073
|
0.483
|
NM_032862
|
TIGD5
|
tigger transposable element derived 5
|
|
chr6_-_43595079
|
0.473
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr6_+_89790412
|
0.472
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr11_-_64684606
|
0.472
|
NM_015104
|
ATG2A
|
ATG2 autophagy related 2 homolog A (S. cerevisiae)
|
|
chr19_-_55628780
|
0.471
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C
|
|
chr6_-_111136616
|
0.471
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr16_-_52581713
|
0.471
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr11_-_89956531
|
0.470
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr8_+_128806720
|
0.468
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr18_+_22040592
|
0.467
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr9_+_131464961
|
0.464
|
|
PKN3
|
protein kinase N3
|
|
chr1_+_110453456
|
0.461
|
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr5_-_64777703
|
0.457
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr19_+_11200037
|
0.456
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr8_-_48650703
|
0.456
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr6_+_138188352
|
0.456
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr6_-_56507585
|
0.455
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr12_-_11150473
|
0.454
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chrX_-_48900932
|
0.454
|
NM_006521
|
TFE3
|
transcription factor binding to IGHM enhancer 3
|
|
chr7_-_16921596
|
0.453
|
NM_176813
|
AGR3
|
anterior gradient 3 homolog (Xenopus laevis)
|
|
chr2_+_28616346
|
0.453
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr10_+_95848811
|
0.448
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr12_+_53443834
|
0.446
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr1_+_43613593
|
0.446
|
NM_001101376
|
FAM183A
|
family with sequence similarity 183, member A
|
|
chr6_+_89790464
|
0.445
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr2_-_145275086
|
0.445
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr20_+_35973083
|
0.443
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr6_-_163148800
|
0.440
|
NM_004562
NM_013987
NM_013988
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr2_+_74756986
|
0.440
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr5_-_172198160
|
0.440
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chrX_-_124097601
|
0.439
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr5_+_71502700
|
0.438
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr17_-_7760740
|
0.436
|
|
LSMD1
|
LSM domain containing 1
|
|
chr12_-_11002051
|
0.433
|
NM_001098538
NM_007244
|
PRR4
|
proline rich 4 (lacrimal)
|
|
chr6_-_33756905
|
0.433
|
NM_181336
|
LEMD2
|
LEM domain containing 2
|
|
chr19_-_40324747
|
0.431
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr12_+_121416548
|
0.430
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr12_+_123868608
|
0.429
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr3_+_115342302
|
0.427
|
|
GAP43
|
growth associated protein 43
|
|
chr6_-_111136330
|
0.427
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr3_-_114866090
|
0.424
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_9268522
|
0.422
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr17_-_80291903
|
0.420
|
NM_003004
|
SECTM1
|
secreted and transmembrane 1
|
|
chr19_-_4831711
|
0.419
|
NM_182919
|
TICAM1
|
toll-like receptor adaptor molecule 1
|
|
chr16_+_69726751
|
0.417
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr12_+_93963597
|
0.417
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr9_-_110252045
|
0.416
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr2_-_74756824
|
0.416
|
|
AUP1
|
ancient ubiquitous protein 1
|