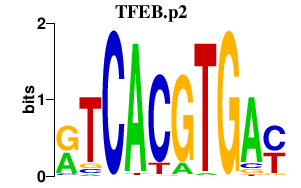
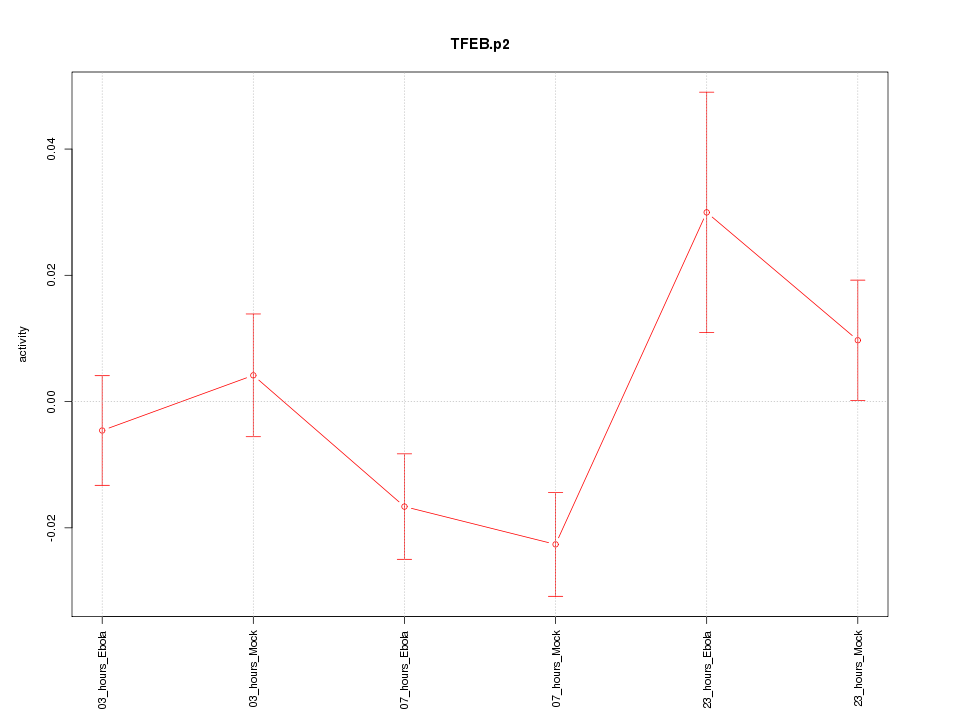
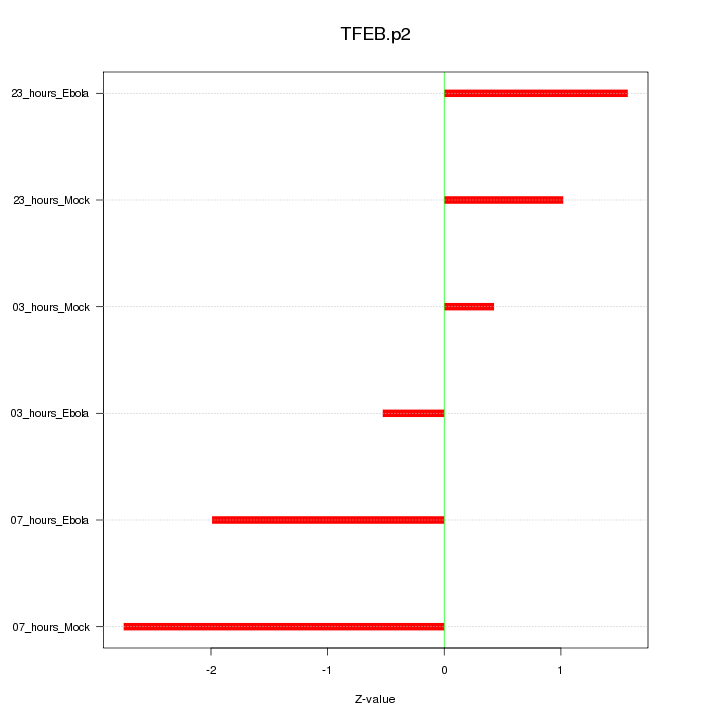
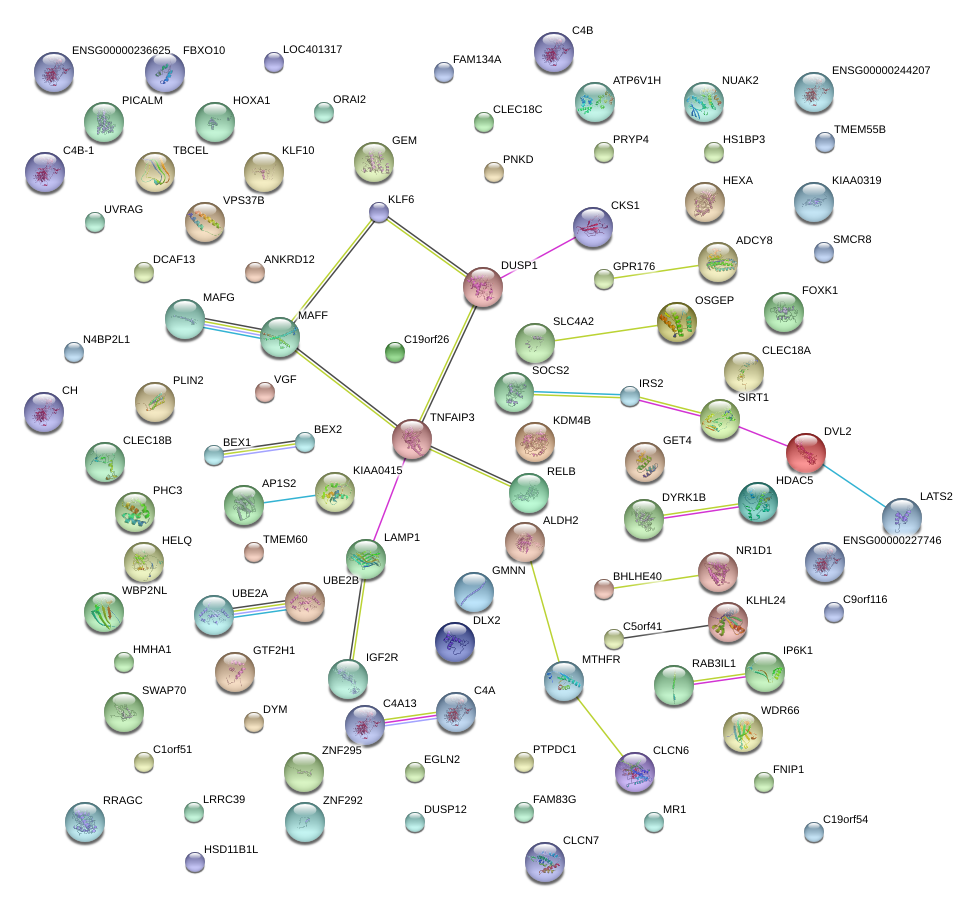
|
chr2_-_172967627
|
3.998
|
|
DLX2
|
distal-less homeobox 2
|
|
chr5_+_172483285
|
2.734
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr2_-_172967234
|
2.356
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr18_+_9136742
|
2.228
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr16_-_1525020
|
2.207
|
|
CLCN7
|
chloride channel 7
|
|
chr1_-_11866079
|
2.091
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr5_-_131132690
|
2.074
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr13_-_110438896
|
2.056
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr5_-_131132642
|
1.993
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr22_+_38599026
|
1.983
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr5_-_131132709
|
1.965
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr16_-_1525006
|
1.957
|
|
CLCN7
|
chloride channel 7
|
|
chr5_-_172198160
|
1.953
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chrX_-_114797017
|
1.917
|
|
|
|
|
chr10_-_3827418
|
1.900
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr16_-_1525040
|
1.872
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr1_-_39325317
|
1.826
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr7_+_27135672
|
1.785
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr19_+_45504686
|
1.762
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr7_-_27135524
|
1.714
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr12_+_122356446
|
1.687
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chrX_-_102319096
|
1.681
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chr1_+_150255228
|
1.673
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr8_-_95274540
|
1.655
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr17_-_7137675
|
1.654
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr5_-_131132715
|
1.645
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr16_-_1524926
|
1.620
|
|
CLCN7
|
chloride channel 7
|
|
chr1_+_11866302
|
1.538
|
|
CLCN6
|
chloride channel 6
|
|
chr17_-_79885544
|
1.513
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr10_-_3827205
|
1.461
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr19_+_5681016
|
1.454
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr7_+_916178
|
1.452
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr8_-_95274520
|
1.375
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr7_+_4815277
|
1.337
|
|
KIAA0415
|
KIAA0415
|
|
chr7_-_100808832
|
1.325
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr22_+_42394728
|
1.308
|
NM_152613
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr17_+_18218569
|
1.241
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr19_+_1067293
|
1.232
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr1_+_11866206
|
1.227
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr7_+_4815258
|
1.185
|
NM_014855
|
KIAA0415
|
KIAA0415
|
|
chr17_-_38256905
|
1.183
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr1_+_11866240
|
1.181
|
|
CLCN6
|
chloride channel 6
|
|
chr7_+_102074002
|
1.176
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr11_-_61684981
|
1.175
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr7_+_28448970
|
1.166
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr13_+_52027435
|
1.156
|
|
|
|
|
chr17_-_18907988
|
1.150
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr1_-_205290756
|
1.146
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr2_-_20850822
|
1.127
|
NM_022460
|
HS1BP3
|
HCLS1 binding protein 3
|
|
chr8_-_54755836
|
1.123
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr3_+_5021141
|
1.123
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr10_+_69644341
|
1.106
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr17_-_7137722
|
1.097
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_-_41256349
|
1.085
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr8_-_54755821
|
1.078
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr17_-_7137580
|
1.078
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr7_-_77427649
|
1.075
|
|
TMEM60
|
transmembrane protein 60
|
|
chr8_-_54755546
|
1.066
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chrX_-_102319080
|
1.056
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr3_+_5021096
|
1.042
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr19_+_5681139
|
1.039
|
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr11_+_18344113
|
1.039
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr13_-_21635629
|
1.036
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr22_-_41593466
|
1.034
|
|
|
|
|
chr16_+_69984809
|
1.014
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr19_-_10764449
|
0.992
|
|
LOC147727
|
uncharacterized LOC147727
|
|
chr8_-_103667882
|
0.983
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr17_+_18218605
|
0.970
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr13_-_21635703
|
0.968
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr6_+_87865261
|
0.952
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr2_-_220042731
|
0.939
|
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr19_-_41256371
|
0.935
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr19_-_1237840
|
0.930
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr12_+_93964801
|
0.924
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr4_-_84376943
|
0.919
|
NM_133636
|
HELQ
|
helicase, POLQ-like
|
|
chr3_+_183353355
|
0.907
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr6_+_31982538
|
0.905
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr6_+_160390128
|
0.901
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr5_+_133706869
|
0.898
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr14_-_20929484
|
0.894
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr11_+_9685624
|
0.894
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr2_+_220042963
|
0.892
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr17_-_38256548
|
0.891
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr17_+_18218608
|
0.890
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr6_+_138188352
|
0.888
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr17_-_42201009
|
0.888
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr13_-_33002218
|
0.888
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr9_+_96793046
|
0.886
|
NM_177995
NM_001253830
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr7_+_150759633
|
0.885
|
NM_001199693
NM_001199694
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr19_+_41305100
|
0.881
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr19_-_40324747
|
0.876
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr17_+_62502860
|
0.876
|
|
CEP95
|
centrosomal protein 95kDa
|
|
chr11_+_9685650
|
0.874
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr17_-_42200957
|
0.859
|
|
HDAC5
|
histone deacetylase 5
|
|
chr19_+_4969123
|
0.858
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr14_-_20929635
|
0.843
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr18_-_46987016
|
0.843
|
NM_017653
|
DYM
|
dymeclin
|
|
chr19_+_41305041
|
0.829
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr15_-_72668184
|
0.827
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr13_+_113951461
|
0.825
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr14_-_20923266
|
0.822
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr15_-_72668301
|
0.812
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr11_+_120894756
|
0.811
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chr21_-_43430448
|
0.810
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr16_+_69984625
|
0.809
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr11_+_9685606
|
0.798
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr3_-_169899519
|
0.796
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr9_-_138391696
|
0.793
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chrX_-_15873094
|
0.791
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr15_-_40213087
|
0.789
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr1_-_100643770
|
0.783
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr11_-_85780088
|
0.782
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr3_-_49823953
|
0.766
|
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr7_+_102073953
|
0.765
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr11_+_75526211
|
0.765
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr19_+_1067536
|
0.758
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr11_+_9685653
|
0.754
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chrX_-_102565805
|
0.750
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr12_-_123380628
|
0.742
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr1_+_181002560
|
0.740
|
NM_001194999
NM_001195000
NM_001195035
NM_001531
|
MR1
|
major histocompatibility complex, class I-related
|
|
chr9_-_19127462
|
0.739
|
|
PLIN2
|
perilipin 2
|
|
chr9_-_37576248
|
0.737
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr1_+_44440642
|
0.732
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_+_45401269
|
0.729
|
NM_001195192
NM_152347
|
C17orf57
|
chromosome 17 open reading frame 57
|
|
chr19_-_7701852
|
0.725
|
|
|
|
|
chr12_-_105501410
|
0.721
|
|
LOC414300
|
uncharacterized LOC414300
|
|
chr7_-_130792839
|
0.719
|
|
FLJ43663
|
uncharacterized LOC378805
|
|
chr12_-_109027654
|
0.718
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr5_+_178977559
|
0.711
|
NM_025158
|
RUFY1
|
RUN and FYVE domain containing 1
|
|
chr11_+_9595309
|
0.711
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr10_-_100206657
|
0.710
|
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr7_-_6312137
|
0.701
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr2_+_239047362
|
0.699
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr17_-_74722843
|
0.699
|
NM_001081461
NM_015167
|
JMJD6
|
jumonji domain containing 6
|
|
chr5_-_132299248
|
0.698
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr19_-_5720155
|
0.694
|
|
LONP1
|
lon peptidase 1, mitochondrial
|
|
chr3_+_5021305
|
0.689
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr11_-_47470687
|
0.687
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr7_+_17338182
|
0.685
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr19_-_10047014
|
0.683
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr19_-_12792698
|
0.682
|
NM_001930
NM_013406
|
DHPS
|
deoxyhypusine synthase
|
|
chr8_+_142402108
|
0.680
|
|
|
|
|
chr3_-_49823972
|
0.677
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr15_-_72410226
|
0.675
|
NM_006901
|
MYO9A
|
myosin IXA
|
|
chr7_-_149157866
|
0.675
|
NM_015694
|
ZNF777
|
zinc finger protein 777
|
|
chr1_+_55681061
|
0.675
|
|
LOC100507634
|
uncharacterized LOC100507634
|
|
chr7_-_100493481
|
0.673
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr22_-_39268214
|
0.671
|
|
CBX6
|
chromobox homolog 6
|
|
chr19_-_5720136
|
0.669
|
|
LONP1
|
lon peptidase 1, mitochondrial
|
|
chrX_+_118108575
|
0.668
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr11_+_118938528
|
0.668
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr14_-_35591439
|
0.663
|
NM_017917
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr10_+_69644708
|
0.663
|
|
SIRT1
|
sirtuin 1
|
|
chr14_-_35591151
|
0.660
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr7_-_27183225
|
0.660
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr5_-_1112106
|
0.650
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr19_-_5720158
|
0.648
|
NM_004793
|
LONP1
|
lon peptidase 1, mitochondrial
|
|
chr1_-_11865963
|
0.648
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr12_-_109027615
|
0.647
|
|
SELPLG
|
selectin P ligand
|
|
chr1_+_218518675
|
0.647
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr1_+_22778343
|
0.646
|
NM_001083621
NM_014870
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr11_+_118938530
|
0.643
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr3_+_51428720
|
0.642
|
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr5_-_132299263
|
0.639
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr22_-_39268169
|
0.638
|
|
CBX6
|
chromobox homolog 6
|
|
chr1_-_145610934
|
0.638
|
|
POLR3C
|
polymerase (RNA) III (DNA directed) polypeptide C (62kD)
|
|
chr14_-_68283293
|
0.636
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr9_-_19127494
|
0.632
|
NM_001122
|
PLIN2
|
perilipin 2
|
|
chr11_-_9286832
|
0.632
|
NM_001243254
NM_015213
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr3_+_51428698
|
0.629
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr11_+_75526236
|
0.628
|
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr11_+_118938515
|
0.628
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr7_-_2354010
|
0.627
|
|
SNX8
|
sorting nexin 8
|
|
chrX_+_100805415
|
0.623
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr11_+_118938473
|
0.622
|
NM_021729
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chrX_+_95939661
|
0.621
|
NM_006729
NM_007309
|
DIAPH2
|
diaphanous homolog 2 (Drosophila)
|
|
chr12_+_96337070
|
0.616
|
NM_152435
|
AMDHD1
|
amidohydrolase domain containing 1
|
|
chr2_+_241526130
|
0.614
|
NM_023083
NM_023085
|
CAPN10
|
calpain 10
|
|
chr1_+_44440623
|
0.609
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr1_-_145610869
|
0.607
|
NM_006468
|
POLR3C
|
polymerase (RNA) III (DNA directed) polypeptide C (62kD)
|
|
chr17_-_44270144
|
0.603
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr11_+_9595183
|
0.602
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr10_-_74385853
|
0.601
|
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr15_-_72668740
|
0.600
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr19_-_10426673
|
0.599
|
NM_001031734
|
FDX1L
|
ferredoxin 1-like
|
|
chr17_+_62503079
|
0.598
|
NM_138363
|
CEP95
|
centrosomal protein 95kDa
|
|
chr14_+_24630421
|
0.596
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr3_-_8693723
|
0.591
|
NM_015931
|
C3orf32
|
chromosome 3 open reading frame 32
|
|
chr11_+_18343794
|
0.591
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr1_+_151170978
|
0.591
|
NM_001135636
NM_001135637
NM_001135638
NM_003557
|
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha
|
|
chr14_+_103388992
|
0.590
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr19_-_12833461
|
0.589
|
NM_001136196
|
TNPO2
|
transportin 2
|
|
chr11_+_9595451
|
0.587
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr14_-_92572950
|
0.584
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr6_-_90121911
|
0.583
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr8_-_8750739
|
0.582
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr10_-_74385818
|
0.578
|
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr7_-_1595746
|
0.575
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr1_+_113498984
|
0.574
|
|
LOC100506392
|
uncharacterized LOC100506392
|
|
chr5_-_72744351
|
0.573
|
NM_004472
|
FOXD1
|
forkhead box D1
|