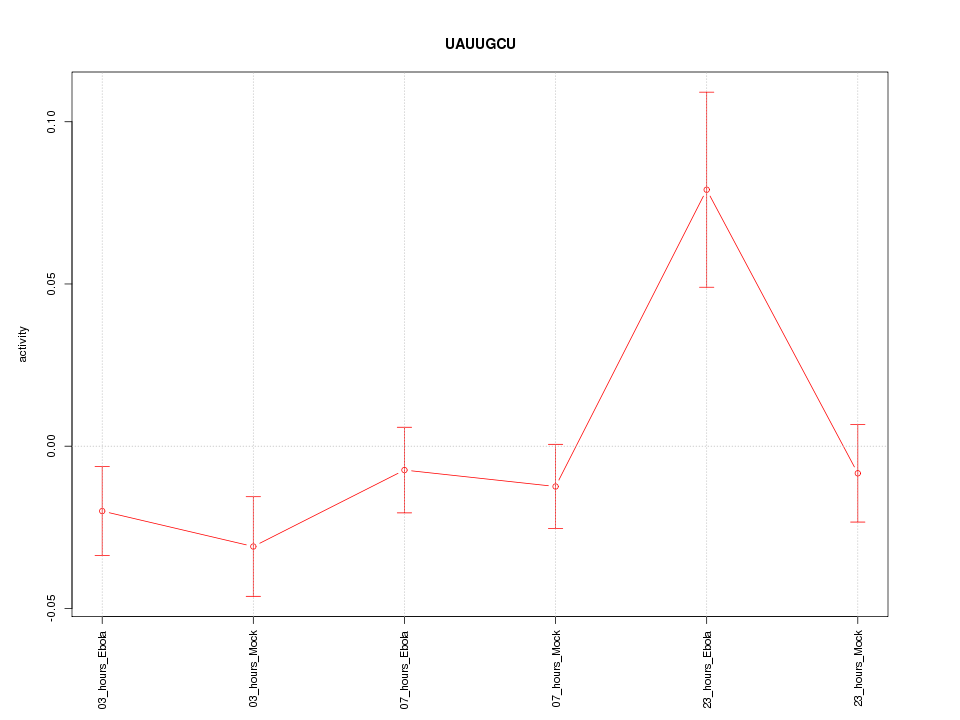
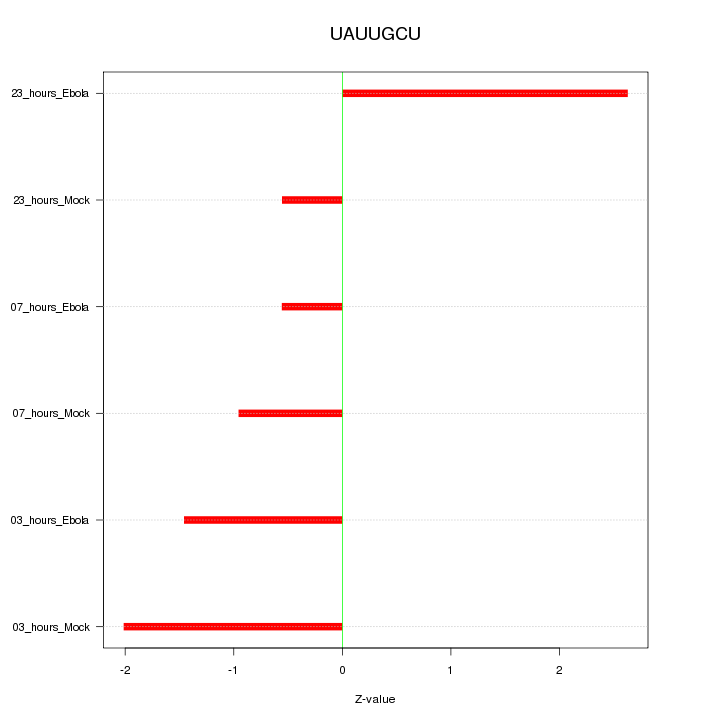
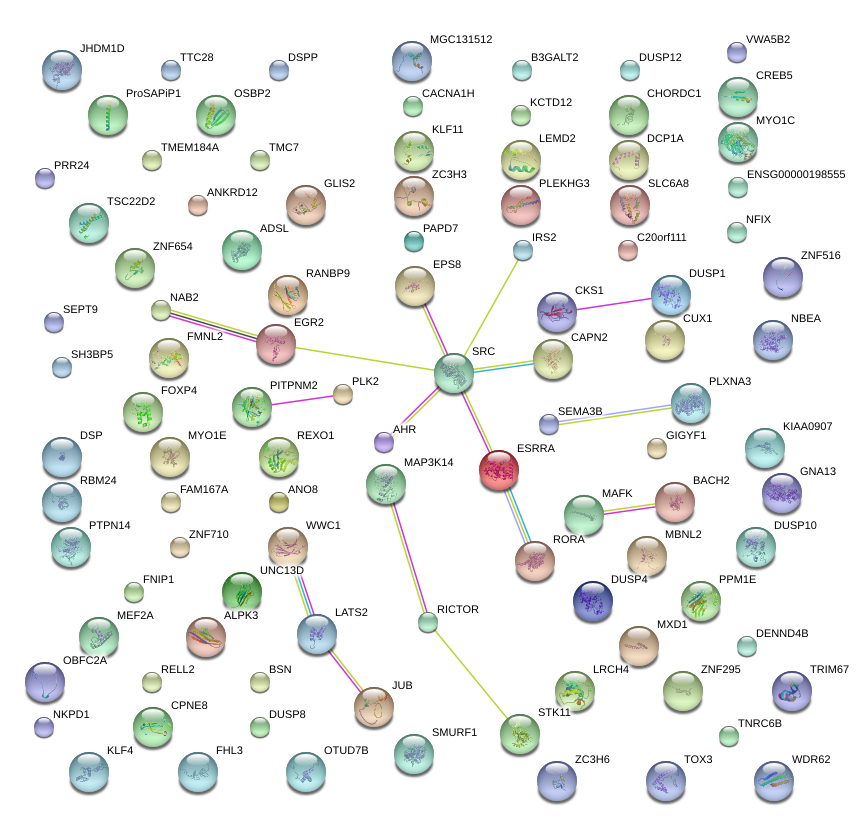
|
chr15_+_85359910
|
3.223
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr13_-_110438896
|
3.021
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr7_+_28475233
|
2.879
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_28338939
|
2.709
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_1570321
|
2.696
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr7_+_28452143
|
2.628
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_-_1593149
|
2.551
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr6_-_91006460
|
2.520
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_-_91006580
|
2.501
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr1_-_38471170
|
2.259
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr6_+_17281773
|
2.089
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr7_+_28725597
|
2.069
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_-_57755787
|
2.064
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr8_-_29206321
|
2.057
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr1_-_193155723
|
2.039
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr15_-_60884615
|
1.728
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr16_+_4382224
|
1.719
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr1_-_221915458
|
1.669
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr13_+_36050885
|
1.651
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr6_+_17282931
|
1.646
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr8_-_11324184
|
1.645
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr15_-_60919642
|
1.643
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_49591897
|
1.639
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr8_-_29207795
|
1.639
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chrX_+_153686620
|
1.633
|
NM_017514
|
PLXNA3
|
plexin A3
|
|
chr1_-_221910801
|
1.616
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr7_-_1595746
|
1.575
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr15_-_61521478
|
1.568
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr6_+_17282294
|
1.549
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr18_+_9137560
|
1.438
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr3_+_50304989
|
1.399
|
NM_001005914
NM_004636
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr15_+_90544608
|
1.395
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chrX_+_152954965
|
1.364
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr9_-_110252045
|
1.336
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr17_-_63052884
|
1.304
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr12_+_57482674
|
1.298
|
NM_005967
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr5_-_172198160
|
1.290
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr7_+_101460881
|
1.260
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr3_+_88188261
|
1.254
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr16_-_52581713
|
1.236
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr8_-_144623611
|
1.217
|
NM_015117
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr19_+_13105970
|
1.186
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr3_+_183948216
|
1.186
|
NM_138345
|
VWA5B2
|
von Willebrand factor A domain containing 5B2
|
|
chr14_+_65171149
|
1.184
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr16_+_18995255
|
1.183
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr12_-_123594974
|
1.177
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr21_-_43430448
|
1.175
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr17_+_56833231
|
1.174
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr12_-_15942350
|
1.173
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chrX_+_152953751
|
1.152
|
NM_001142805
NM_005629
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr7_-_98741681
|
1.151
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr1_-_214724974
|
1.150
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr16_+_18995508
|
1.147
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr3_-_15382874
|
1.141
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr12_-_39299335
|
1.130
|
NM_153634
|
CPNE8
|
copine VIII
|
|
chr3_-_15374082
|
1.103
|
NM_004844
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr7_-_100286869
|
1.063
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr6_+_7541863
|
1.044
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr19_-_17445538
|
1.037
|
NM_020959
|
ANO8
|
anoctamin 8
|
|
chr16_+_1203240
|
1.037
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr16_-_52580805
|
1.034
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr5_-_131132715
|
1.034
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr13_+_97874540
|
1.033
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr19_+_47778099
|
1.032
|
NM_178511
|
PRR24
|
proline rich 24
|
|
chr14_-_23451704
|
1.029
|
NM_032876
|
AJUBA
|
ajuba LIM protein
|
|
chr17_-_43394326
|
1.028
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr22_+_40573879
|
1.010
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_-_155904186
|
1.002
|
NM_014949
|
KIAA0907
|
KIAA0907
|
|
chr22_-_29075708
|
0.980
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr2_+_10184371
|
0.974
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr1_+_223900061
|
0.970
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr19_-_1848447
|
0.962
|
NM_020695
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)
|
|
chr20_+_35973083
|
0.955
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr18_-_74207145
|
0.954
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr5_+_141016516
|
0.945
|
NM_001130029
NM_173828
|
RELL2
|
RELT-like 2
|
|
chr5_-_39074494
|
0.932
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr6_+_41514133
|
0.929
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr22_+_31090792
|
0.927
|
NM_030758
|
OSBP2
|
oxysterol binding protein 2
|
|
chr3_-_53381533
|
0.921
|
NM_018403
|
DCP1A
|
DCP1 decapping enzyme homolog A (S. cerevisiae)
|
|
chr17_-_1394849
|
0.912
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr13_-_77460432
|
0.906
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr18_+_9136742
|
0.905
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr19_+_1205773
|
0.896
|
NM_000455
|
STK11
|
serine/threonine kinase 11
|
|
chr10_-_64578926
|
0.895
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr13_+_35516390
|
0.893
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr2_+_192542860
|
0.885
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr2_+_113033146
|
0.884
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr15_+_100106132
|
0.880
|
NM_001130926
NM_001130927
NM_001171894
NM_005587
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr2_+_153191591
|
0.878
|
NM_052905
|
FMNL2
|
formin-like 2
|
|
chr2_+_70142152
|
0.877
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr7_+_17338182
|
0.875
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr11_-_89956531
|
0.869
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr11_+_64073040
|
0.867
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr20_-_3149070
|
0.857
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr3_+_150126705
|
0.849
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr5_+_167718485
|
0.846
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr20_-_42839423
|
0.842
|
NM_016470
|
C20orf111
|
chromosome 20 open reading frame 111
|
|
chr13_-_21635703
|
0.839
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr7_-_100183753
|
0.838
|
NM_002319
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr5_+_6714717
|
0.836
|
NM_001171805
NM_006999
|
PAPD7
|
PAP associated domain containing 7
|
|
chr1_+_231298673
|
0.834
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr7_-_139876718
|
0.828
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr5_+_6746100
|
0.813
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr6_-_13711643
|
0.787
|
NM_005493
|
RANBP9
|
RAN binding protein 9
|
|
chr6_-_33754712
|
0.786
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr1_-_153919140
|
0.785
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr15_+_100173182
|
0.783
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr1_-_149982528
|
0.780
|
NM_020205
|
OTUD7B
|
OTU domain containing 7B
|
|
chr19_-_45663407
|
0.779
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr1_+_154975041
|
0.771
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr17_-_1395950
|
0.771
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr1_+_95558072
|
0.764
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr15_+_57210823
|
0.758
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr7_-_35734762
|
0.754
|
NM_022373
|
HERPUD2
|
HERPUD family member 2
|
|
chr6_-_33756905
|
0.744
|
NM_181336
|
LEMD2
|
LEM domain containing 2
|
|
chr2_-_158454025
|
0.731
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr19_-_4066739
|
0.730
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr12_-_54121203
|
0.728
|
NM_001143682
NM_020898
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr19_-_2721337
|
0.728
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr2_+_10183630
|
0.724
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr12_+_110437233
|
0.720
|
NM_033121
|
ANKRD13A
|
ankyrin repeat domain 13A
|
|
chr8_+_21946636
|
0.717
|
NM_022749
|
FAM160B2
|
family with sequence similarity 160, member B2
|
|
chr1_+_223889253
|
0.714
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_+_236305831
|
0.712
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr16_-_67450149
|
0.709
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr4_+_108814419
|
0.708
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr5_+_78531848
|
0.703
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr22_-_50218451
|
0.701
|
NM_014577
|
BRD1
|
bromodomain containing 1
|
|
chr11_-_63684315
|
0.693
|
NM_173587
|
RCOR2
|
REST corepressor 2
|
|
chr22_+_25423940
|
0.693
|
NM_001145206
|
KIAA1671
|
KIAA1671
|
|
chr6_-_111136407
|
0.688
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr4_-_153303663
|
0.688
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr15_+_57511616
|
0.685
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr4_-_184241926
|
0.685
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr20_+_35974458
|
0.682
|
NM_198291
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr11_-_70935807
|
0.682
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr10_-_94050799
|
0.672
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr18_-_21166440
|
0.669
|
NM_000271
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr1_+_36273786
|
0.667
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr7_-_22396511
|
0.663
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr12_+_8850479
|
0.661
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr22_+_40440664
|
0.656
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr20_-_32262254
|
0.644
|
NM_031231
NM_031232
|
NECAB3
|
N-terminal EF-hand calcium binding protein 3
|
|
chr3_-_24536176
|
0.631
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr17_-_27169740
|
0.629
|
NM_001077498
NM_018182
|
C17orf63
|
chromosome 17 open reading frame 63
|
|
chr14_+_62229062
|
0.628
|
NM_003082
|
SNAPC1
|
small nuclear RNA activating complex, polypeptide 1, 43kDa
|
|
chr1_+_95582827
|
0.627
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr11_+_92085261
|
0.624
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr12_+_79439432
|
0.622
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr19_+_45596406
|
0.621
|
NM_019121
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37
|
|
chr20_+_48599511
|
0.617
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chrX_+_118108595
|
0.617
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr6_+_42788793
|
0.616
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr3_+_141043054
|
0.612
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr12_+_58148775
|
0.609
|
NM_138396
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9
|
|
chr16_-_402617
|
0.609
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chrX_+_13707236
|
0.604
|
NM_001195328
NM_004251
|
RAB9A
|
RAB9A, member RAS oncogene family
|
|
chr6_-_31763551
|
0.603
|
NM_006295
|
VARS
|
valyl-tRNA synthetase
|
|
chr1_-_155532322
|
0.601
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr19_+_7459964
|
0.601
|
NM_015318
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr1_+_185126191
|
0.601
|
NM_001105518
NM_017673
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae)
|
|
chr12_+_79257772
|
0.597
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr11_+_47430045
|
0.594
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr7_-_44105162
|
0.594
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr2_+_149402559
|
0.591
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr1_-_205180691
|
0.591
|
NM_015375
NM_199462
|
DSTYK
|
dual serine/threonine and tyrosine protein kinase
|
|
chr17_-_1389013
|
0.587
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr4_-_69215699
|
0.584
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr6_-_37467688
|
0.584
|
NM_138493
|
CCDC167
|
coiled-coil domain containing 167
|
|
chr17_-_7387513
|
0.582
|
NM_020899
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr14_-_23446431
|
0.581
|
NM_198086
|
AJUBA
|
ajuba LIM protein
|
|
chrX_-_48828250
|
0.580
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr19_+_7504573
|
0.575
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr2_+_192109879
|
0.573
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr14_-_24911658
|
0.572
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr6_-_24383519
|
0.569
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr11_-_790103
|
0.567
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr4_+_108745718
|
0.566
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr5_+_149887673
|
0.565
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr4_-_186456580
|
0.561
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr8_-_52811707
|
0.560
|
NM_052937
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr12_-_31743822
|
0.558
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr7_-_98030426
|
0.556
|
NM_018842
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr9_-_138987096
|
0.550
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr22_+_47022657
|
0.549
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr10_+_98592658
|
0.544
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr4_-_66535652
|
0.538
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr5_+_49961732
|
0.538
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr18_+_29671817
|
0.533
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr17_-_1532109
|
0.533
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr12_-_49504646
|
0.531
|
NM_018113
|
LMBR1L
|
limb region 1 homolog (mouse)-like
|
|
chr10_-_94003002
|
0.531
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr2_+_173940505
|
0.529
|
NM_016653
NM_133646
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr21_-_19191643
|
0.518
|
NM_001100420
NM_001100421
NM_017447
|
C21orf91
|
chromosome 21 open reading frame 91
|
|
chr10_-_735522
|
0.516
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr2_-_158485393
|
0.512
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr4_+_186131283
|
0.509
|
NM_031953
|
SNX25
|
sorting nexin 25
|
|
chr15_+_44092618
|
0.506
|
NM_001199885
NM_016400
|
C15orf63
|
chromosome 15 open reading frame 63
|
|
chr17_+_1959512
|
0.501
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr5_+_125800786
|
0.494
|
NM_001146322
|
GRAMD3
|
GRAM domain containing 3
|