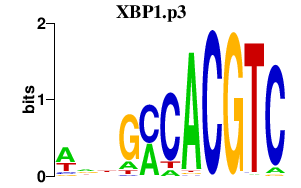
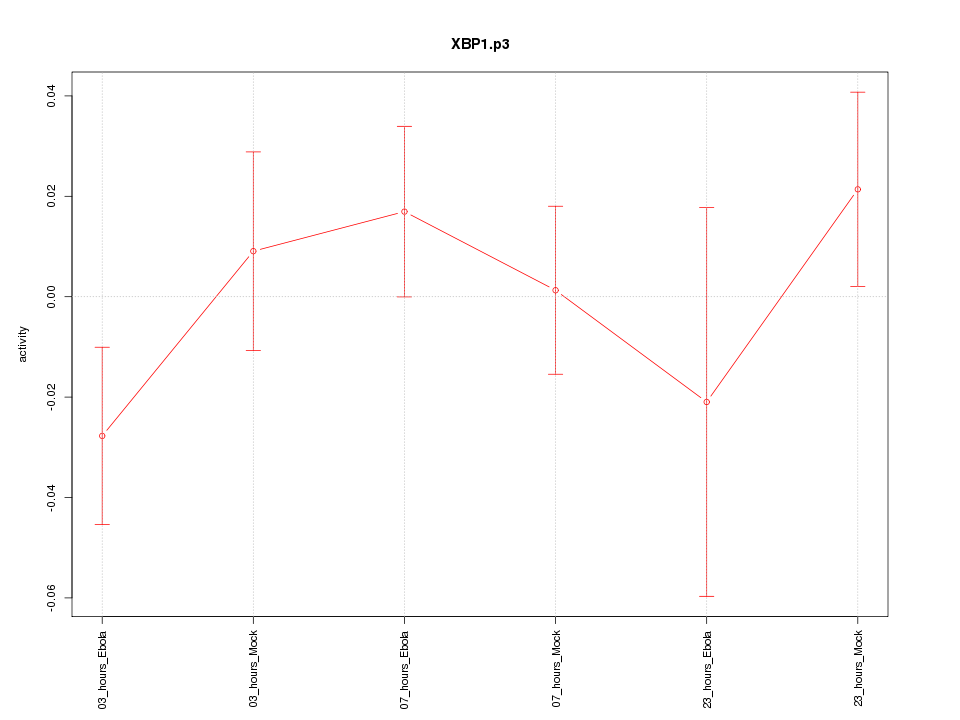
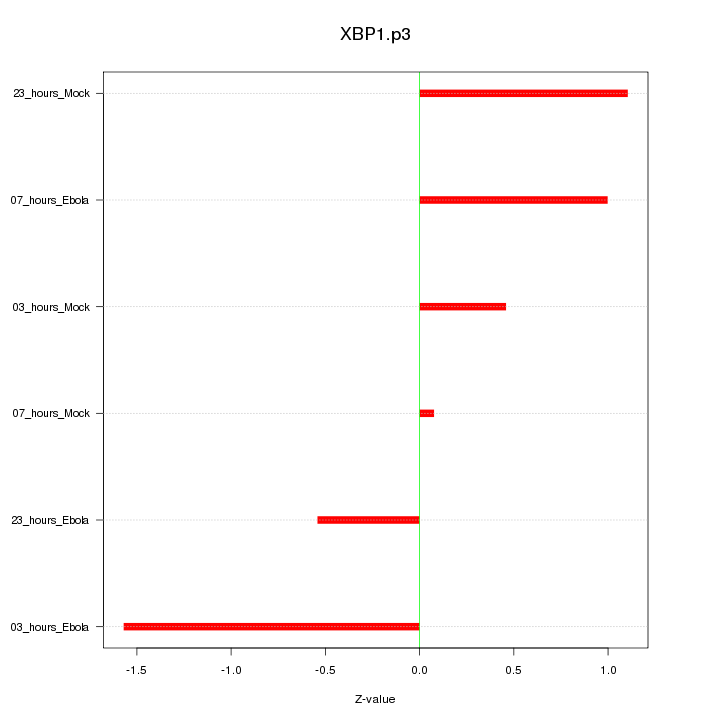
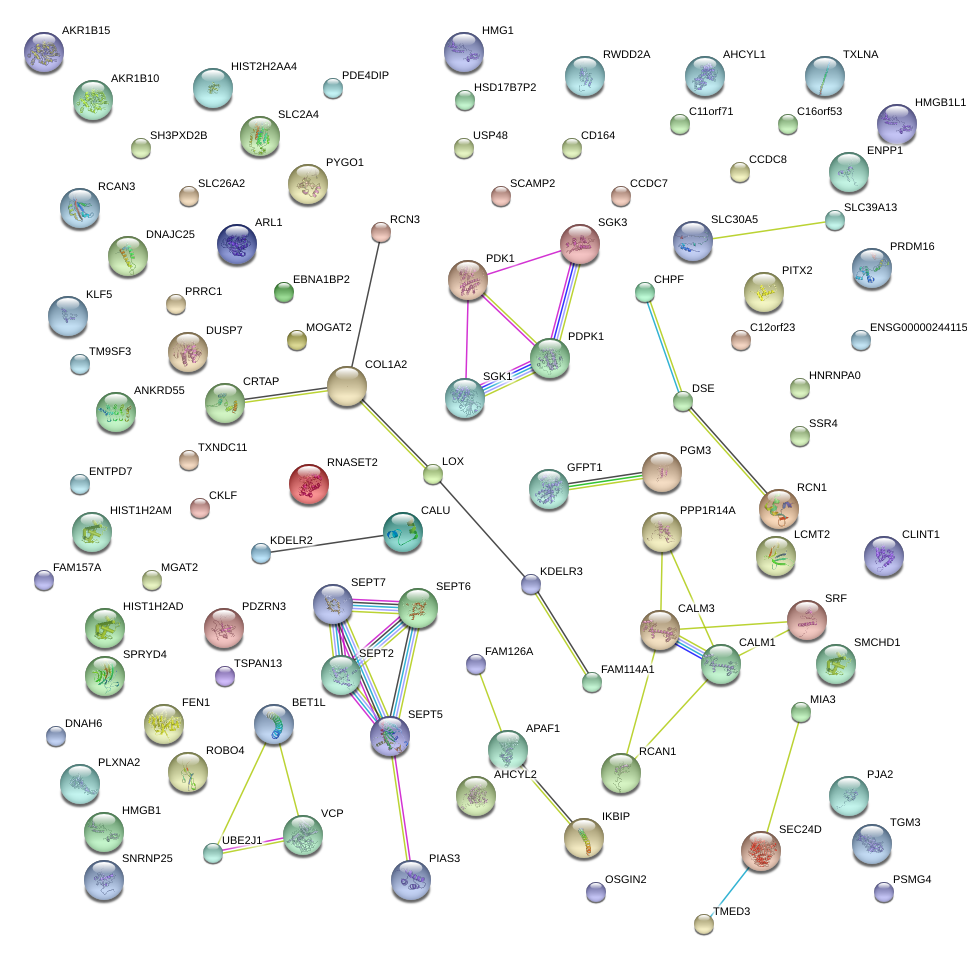
|
chr12_-_99038401
|
0.770
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chr19_+_50031232
|
0.549
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr15_-_55880508
|
0.451
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr15_-_55881049
|
0.435
|
NM_015617
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr1_-_144931967
|
0.397
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr7_-_6523689
|
0.388
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr17_+_34890796
|
0.370
|
|
|
|
|
chr19_-_38746978
|
0.358
|
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr14_+_50087400
|
0.357
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr5_-_121413906
|
0.351
|
|
LOX
|
lysyl oxidase
|
|
chr4_-_76649557
|
0.328
|
|
|
|
|
chr19_+_50030874
|
0.313
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr5_-_108745569
|
0.308
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr1_+_119683006
|
0.296
|
|
|
|
|
chr5_-_121414011
|
0.278
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr12_+_56862300
|
0.276
|
NM_207344
|
SPRYD4
|
SPRY domain containing 4
|
|
chr11_+_61560352
|
0.274
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr4_+_38869346
|
0.261
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr22_+_38864015
|
0.260
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr6_+_3259134
|
0.259
|
NM_001128591
NM_001128592
NM_001135750
|
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4
|
|
chr7_+_16793320
|
0.257
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr5_+_126853308
|
0.257
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr11_-_114271052
|
0.256
|
NM_019021
|
C11orf71
|
chromosome 11 open reading frame 71
|
|
chr11_+_61560108
|
0.254
|
NM_004111
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr11_+_61560401
|
0.246
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr3_-_52090090
|
0.245
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr15_-_43622687
|
0.243
|
NM_014793
|
LCMT2
|
leucine carboxyl methyltransferase 2
|
|
chr16_-_75529278
|
0.236
|
|
|
|
|
chr6_-_83902921
|
0.235
|
NM_001199918
NM_001199919
NM_015599
|
PGM3
|
phosphoglucomutase 3
|
|
chr7_+_128379395
|
0.231
|
|
CALU
|
calumenin
|
|
chr2_+_84743576
|
0.223
|
NM_001370
|
DNAH6
|
dynein, axonemal, heavy chain 6
|
|
chr7_+_128379428
|
0.217
|
|
CALU
|
calumenin
|
|
chr13_-_31040035
|
0.214
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr5_-_137089754
|
0.212
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr7_+_128379400
|
0.211
|
|
CALU
|
calumenin
|
|
chr5_+_68389856
|
0.206
|
|
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5
|
|
chr6_+_116692098
|
0.205
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr2_+_173420794
|
0.203
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr13_-_31039966
|
0.202
|
|
HMGB1
|
high mobility group box 1
|
|
chr12_-_99038705
|
0.201
|
|
IKBIP
|
IKBKB interacting protein
|
|
chr7_+_128379452
|
0.200
|
|
CALU
|
calumenin
|
|
chr3_-_73673978
|
0.196
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr5_-_137089434
|
0.193
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr10_-_98346766
|
0.186
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr17_+_7184978
|
0.186
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr7_+_128379345
|
0.185
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr7_-_23053732
|
0.183
|
NM_032581
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr19_-_39522867
|
0.181
|
|
|
|
|
chr11_+_32112607
|
0.179
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr12_+_107349673
|
0.178
|
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr5_-_137088952
|
0.176
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr19_-_46916825
|
0.175
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr7_+_94023872
|
0.173
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr10_-_98346793
|
0.172
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr7_-_23053674
|
0.170
|
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr2_-_69614321
|
0.170
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr19_-_50354305
|
0.167
|
|
LOC100506033
|
uncharacterized LOC100506033
|
|
chr4_-_111544206
|
0.167
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr2_-_220408264
|
0.167
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr6_+_27100784
|
0.165
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr4_-_119757183
|
0.164
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr5_+_68389911
|
0.163
|
|
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5
|
|
chr11_-_207391
|
0.162
|
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr1_+_222791151
|
0.160
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr16_+_66586447
|
0.160
|
NM_001202509
NM_001204098
NM_001204099
NM_001040138
NM_016326
NM_016951
NM_181640
NM_181641
|
CKLF-CMTM1
CKLF
|
CKLF-CMTM1 readthrough
chemokine-like factor
|
|
chr11_+_47430045
|
0.157
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr6_-_109703553
|
0.157
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr1_+_145576011
|
0.156
|
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr18_-_34409084
|
0.153
|
NM_015476
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr6_+_132129154
|
0.153
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr5_-_121413967
|
0.153
|
|
LOX
|
lysyl oxidase
|
|
chr5_+_149340346
|
0.151
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr18_-_34408845
|
0.151
|
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr16_-_75529289
|
0.150
|
|
|
|
|
chr2_+_173420776
|
0.149
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr1_-_208417635
|
0.148
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr16_+_29827489
|
0.148
|
NM_024516
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr15_-_75165595
|
0.147
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr18_-_34408948
|
0.147
|
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr10_-_98346594
|
0.146
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_+_83903031
|
0.143
|
NM_033411
|
RWDD2A
|
RWD domain containing 2A
|
|
chr1_+_110527327
|
0.142
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr5_+_68389775
|
0.141
|
NM_001251969
NM_022902
NM_024055
|
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5
|
|
chr1_-_145039631
|
0.141
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr5_+_68389916
|
0.140
|
|
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5
|
|
chr5_-_157285977
|
0.137
|
NM_001195555
NM_001195556
NM_014666
|
CLINT1
|
clathrin interactor 1
|
|
chr15_+_79603427
|
0.136
|
NM_007364
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr16_+_103892
|
0.135
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr15_+_79603544
|
0.134
|
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr1_-_22109605
|
0.131
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr8_+_67624652
|
0.129
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr19_+_47104389
|
0.129
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr6_-_109702902
|
0.128
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr5_-_171881421
|
0.128
|
NM_001017995
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr12_-_101801499
|
0.128
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr12_+_99038918
|
0.127
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr6_+_132129195
|
0.126
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr11_+_32112414
|
0.125
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr9_+_114393543
|
0.123
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr1_-_43736600
|
0.123
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chrX_+_153060073
|
0.122
|
|
SSR4
|
signal sequence receptor, delta
|
|
chr6_-_167370037
|
0.122
|
NM_003730
|
RNASET2
|
ribonuclease T2
|
|
chr6_-_134496833
|
0.121
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_+_242254722
|
0.121
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr4_-_119757288
|
0.116
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr10_+_101419169
|
0.116
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr7_+_94024201
|
0.115
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr6_-_90062349
|
0.115
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr2_+_173420873
|
0.115
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chrX_+_153059903
|
0.114
|
NM_001204526
NM_006280
|
SSR4
|
signal sequence receptor, delta
|
|
chr9_-_35072545
|
0.113
|
|
VCP
|
valosin containing protein
|
|
chr16_+_66586479
|
0.113
|
|
CKLF
|
chemokine-like factor
|
|
chr1_+_32645647
|
0.112
|
|
TXLNA
|
taxilin alpha
|
|
chr10_+_32735021
|
0.111
|
NM_001026383
NM_145023
|
CCDC7
|
coiled-coil domain containing 7
|
|
chr3_+_33155447
|
0.109
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chr16_-_11836602
|
0.108
|
NM_015914
|
TXNDC11
|
thioredoxin domain containing 11
|
|
chr17_+_55333843
|
0.108
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr1_-_22109669
|
0.107
|
NM_001032730
NM_032236
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr3_+_132379204
|
0.106
|
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr1_-_22109450
|
0.105
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr8_-_81083660
|
0.105
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr9_-_35072596
|
0.103
|
|
VCP
|
valosin containing protein
|
|
chr12_-_133405197
|
0.103
|
NM_005895
|
GOLGA3
|
golgin A3
|
|
chr19_-_47104456
|
0.103
|
NM_001205281
|
LOC100506012
|
serine/threonine-protein phosphatase 5-like
|
|
chr15_-_52030162
|
0.103
|
NM_153374
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2
|
|
chr5_-_171881355
|
0.102
|
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr2_+_136499141
|
0.101
|
NM_014607
|
UBXN4
|
UBX domain protein 4
|
|
chr5_+_149340287
|
0.100
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr9_+_119449573
|
0.100
|
NM_001099679
NM_012210
|
TRIM32
|
tripartite motif containing 32
|
|
chr6_-_106773529
|
0.099
|
|
ATG5
|
ATG5 autophagy related 5 homolog (S. cerevisiae)
|
|
chr7_-_27224762
|
0.098
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr2_+_220408352
|
0.097
|
|
TMEM198
|
transmembrane protein 198
|
|
chr3_-_141944296
|
0.097
|
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr3_-_56717132
|
0.097
|
NM_001112736
|
FAM208A
|
family with sequence similarity 208, member A
|
|
chr18_+_77659446
|
0.096
|
|
|
|
|
chr6_-_90062439
|
0.095
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr12_-_121711980
|
0.095
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr13_-_45915203
|
0.095
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr19_+_47104559
|
0.094
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr15_-_55700456
|
0.093
|
NM_004748
NM_001204450
NM_001204451
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr1_+_110527418
|
0.093
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_+_5904842
|
0.092
|
NM_001017921
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein
|
|
chr10_-_98346558
|
0.091
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr2_-_39187477
|
0.091
|
|
LOC375196
|
uncharacterized LOC375196
|
|
chr13_-_45915308
|
0.091
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr5_+_43603287
|
0.091
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr5_-_101834548
|
0.091
|
NM_173488
|
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1
|
|
chr2_+_183580960
|
0.090
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr20_+_31407696
|
0.090
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr6_+_33168602
|
0.090
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr7_-_128045995
|
0.089
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr4_+_56262051
|
0.089
|
NM_018475
|
TMEM165
|
transmembrane protein 165
|
|
chr5_+_43603216
|
0.089
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr6_-_108279409
|
0.088
|
NM_007214
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr4_-_39460058
|
0.087
|
NM_001024921
|
RPL9
|
ribosomal protein L9
|
|
chr1_+_110527432
|
0.086
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_+_1491000
|
0.085
|
NM_138393
|
REEP6
|
receptor accessory protein 6
|
|
chr12_-_133405403
|
0.084
|
NM_001172557
|
GOLGA3
|
golgin A3
|
|
chr3_+_33155559
|
0.084
|
|
CRTAP
|
cartilage associated protein
|
|
chr19_+_47104490
|
0.084
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr16_+_29827707
|
0.083
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr10_-_71930185
|
0.082
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr10_+_89419352
|
0.082
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr17_+_8339135
|
0.081
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chr7_+_128095881
|
0.081
|
NM_001098786
NM_013332
|
HILPDA
|
hypoxia inducible lipid droplet-associated
|
|
chrX_-_118827322
|
0.080
|
NM_015129
NM_145799
NM_145800
NM_145802
|
SEPT6
|
septin 6
|
|
chr1_-_11120076
|
0.080
|
|
SRM
|
spermidine synthase
|
|
chr5_+_115177616
|
0.079
|
NM_001284
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit
|
|
chr5_-_114938060
|
0.079
|
NM_021649
|
TICAM2
|
toll-like receptor adaptor molecule 2
|
|
chr5_-_60458228
|
0.078
|
NM_001048249
|
C5orf43
|
chromosome 5 open reading frame 43
|
|
chr8_+_6565870
|
0.078
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr5_+_43603312
|
0.078
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr1_-_11120001
|
0.077
|
|
SRM
|
spermidine synthase
|
|
chr5_+_149340437
|
0.077
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr5_+_34656367
|
0.076
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr14_-_39572278
|
0.075
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr21_-_44299660
|
0.075
|
NM_018669
NM_033661
|
WDR4
|
WD repeat domain 4
|
|
chr11_-_31825593
|
0.074
|
|
PAX6
|
paired box 6
|
|
chr20_-_8000370
|
0.074
|
NM_021156
|
TMX4
|
thioredoxin-related transmembrane protein 4
|
|
chr3_+_171758289
|
0.074
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr1_+_145575987
|
0.073
|
NM_006099
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chrX_+_153059629
|
0.073
|
NM_001204527
|
SSR4
|
signal sequence receptor, delta
|
|
chr6_-_90062610
|
0.072
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr9_+_139377946
|
0.072
|
NM_152571
|
C9orf163
|
chromosome 9 open reading frame 163
|
|
chr11_-_207403
|
0.071
|
NM_001098787
NM_016526
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr12_+_89744714
|
0.071
|
|
LOC100131490
|
uncharacterized LOC100131490
|
|
chr9_-_35072643
|
0.070
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr18_-_34408699
|
0.070
|
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr11_-_64851556
|
0.070
|
NM_080668
|
CDCA5
|
cell division cycle associated 5
|
|
chr14_-_24664606
|
0.070
|
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr3_-_57583153
|
0.069
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr2_+_171785624
|
0.069
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr3_-_176914198
|
0.068
|
|
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1
|
|
chr9_-_113800243
|
0.067
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr6_-_90062289
|
0.067
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr10_+_102106771
|
0.067
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr22_-_30685485
|
0.067
|
NM_001037666
|
GATSL3
|
GATS protein-like 3
|
|
chr17_+_14204326
|
0.067
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr15_+_76135621
|
0.066
|
NM_173469
|
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2
|
|
chr11_+_69455938
|
0.066
|
|
CCND1
|
cyclin D1
|