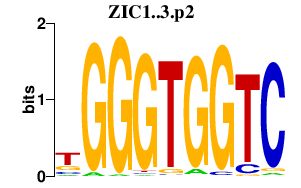
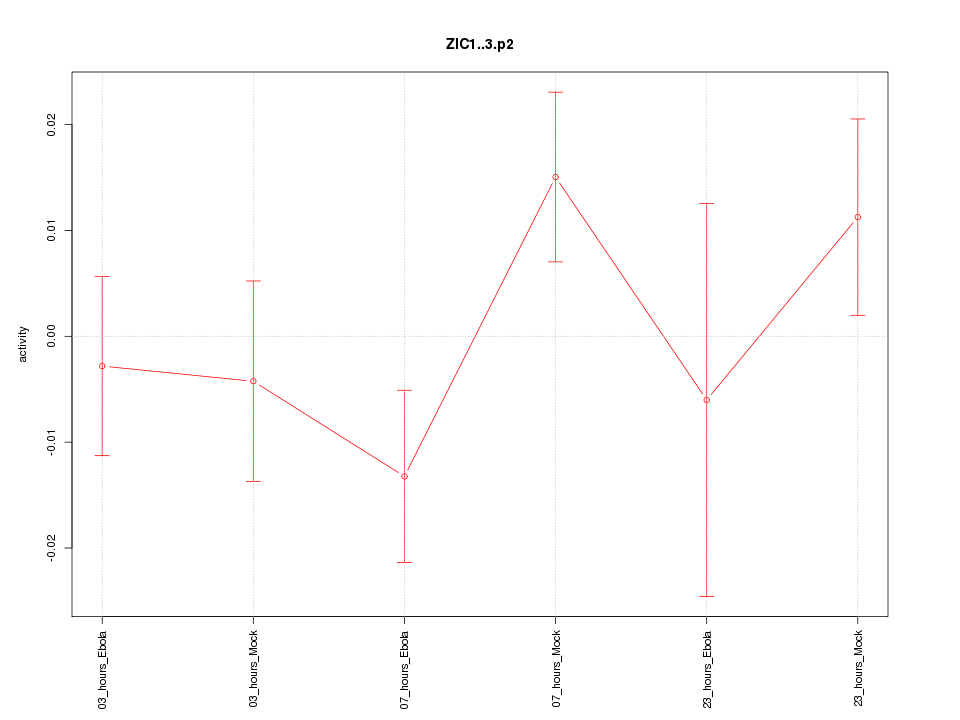
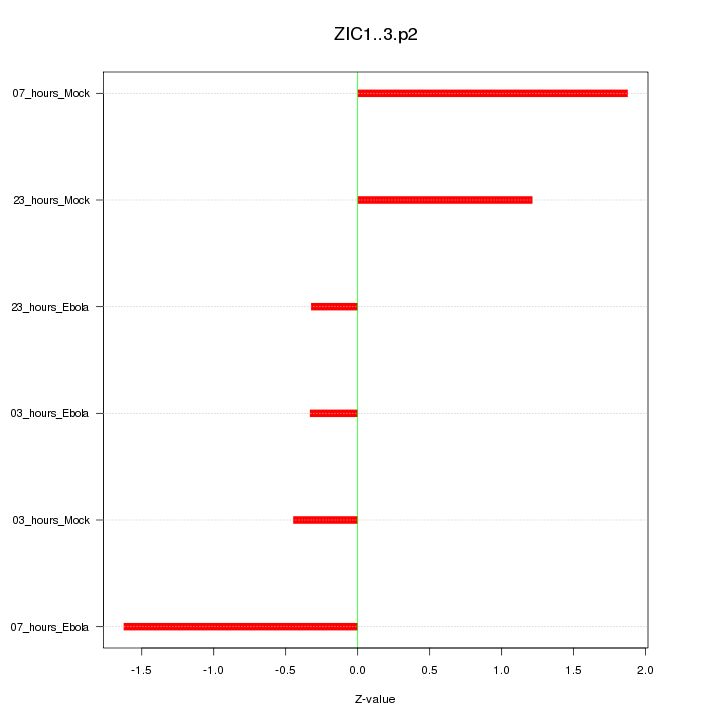
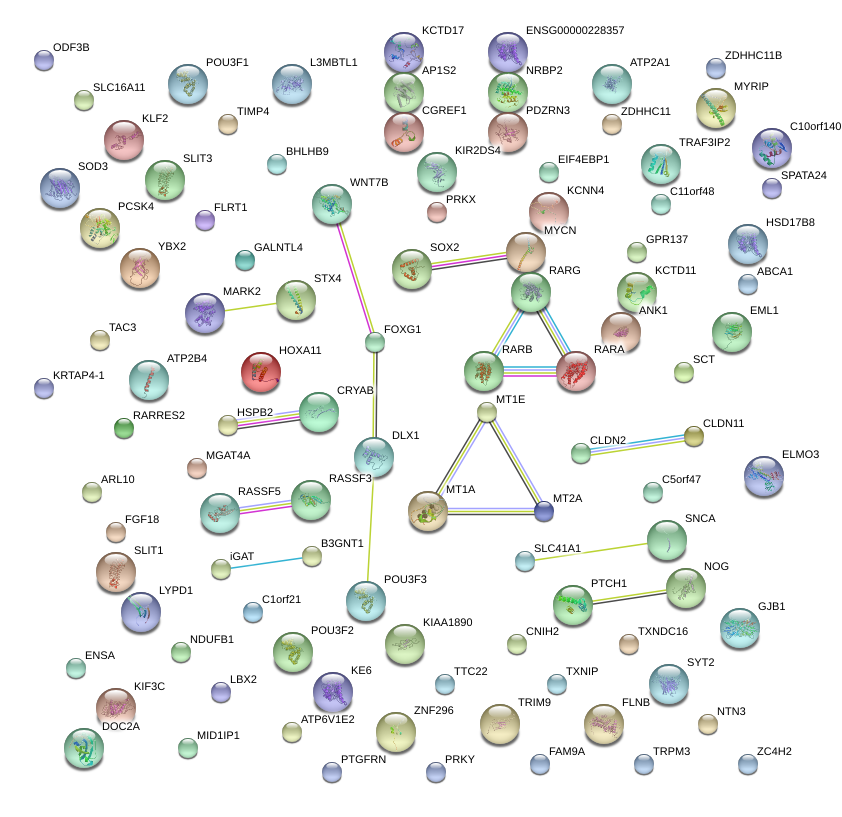
|
chr11_-_111782447
|
0.660
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr3_-_73673978
|
0.620
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr19_+_16435632
|
0.613
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr2_+_16081945
|
0.508
|
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr2_+_16082148
|
0.493
|
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr12_-_57410275
|
0.470
|
NM_001178054
NM_013251
|
TAC3
|
tachykinin 3
|
|
chr7_-_27224762
|
0.465
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr1_-_150602020
|
0.449
|
|
ENSA
|
endosulfine alpha
|
|
chr6_+_19838612
|
0.446
|
|
|
|
|
chr7_+_150102376
|
0.340
|
|
|
|
|
chr20_-_35201677
|
0.330
|
|
|
|
|
chr6_+_33172418
|
0.328
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr14_+_29236882
|
0.300
|
|
FOXG1
|
forkhead box G1
|
|
chr17_-_7197811
|
0.295
|
|
YBX2
|
Y box binding protein 2
|
|
chr9_-_73483933
|
0.288
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr16_+_67233013
|
0.285
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr6_+_99282597
|
0.284
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr2_-_26205327
|
0.280
|
|
KIF3C
|
kinesin family member 3C
|
|
chrX_-_15872921
|
0.262
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr5_+_175792444
|
0.253
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr19_+_7694658
|
0.251
|
NM_001171155
|
C19orf79
|
chromosome 19 open reading frame 79
|
|
chr19_-_45579687
|
0.250
|
NM_145288
|
ZNF296
|
zinc finger protein 296
|
|
chr19_+_16435734
|
0.249
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr7_-_150038689
|
0.245
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr9_+_131870105
|
0.244
|
|
|
|
|
chrX_+_70443055
|
0.241
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr2_-_178257380
|
0.241
|
|
LOC100130691
|
uncharacterized LOC100130691
|
|
chr11_+_66045645
|
0.241
|
NM_182553
|
CNIH2
|
cornichon homolog 2 (Drosophila)
|
|
chr9_-_107666046
|
0.239
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr16_+_56672577
|
0.237
|
NM_005946
|
MT1A
|
metallothionein 1A
|
|
chr5_-_138739755
|
0.235
|
NM_194296
|
SPATA24
|
spermatogenesis associated 24
|
|
chr17_-_39341126
|
0.234
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr5_-_851100
|
0.233
|
NM_024786
|
ZDHHC11
|
zinc finger, DHHC-type containing 11
|
|
chr2_-_27333963
|
0.232
|
NM_001166241
|
CGREF1
|
cell growth regulator with EF-hand domain 1
|
|
chr11_-_62439085
|
0.230
|
|
C11orf48
|
chromosome 11 open reading frame 48
|
|
chrX_+_70443459
|
0.227
|
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr6_+_75994713
|
0.222
|
|
LOC100506804
|
uncharacterized LOC100506804
|
|
chrX_+_106171328
|
0.218
|
|
CLDN2
|
claudin 2
|
|
chr10_-_21805715
|
0.216
|
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr8_-_144923019
|
0.204
|
NM_178564
|
NRBP2
|
nuclear receptor binding protein 2
|
|
chr5_-_168727715
|
0.203
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr6_-_111927473
|
0.202
|
NM_001164281
NM_147686
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr3_-_12200850
|
0.200
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr11_+_63871240
|
0.199
|
NM_013280
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr1_-_205782135
|
0.198
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr12_-_53613883
|
0.197
|
|
RARG
|
retinoic acid receptor, gamma
|
|
chr15_-_23378167
|
0.196
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr5_+_170847059
|
0.194
|
|
FGF18
|
fibroblast growth factor 18
|
|
chr9_-_98278939
|
0.192
|
|
PTCH1
|
patched 1
|
|
chrX_-_64196268
|
0.192
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chrX_-_15872905
|
0.189
|
|
|
|
|
chr6_-_111927059
|
0.185
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr3_+_170136652
|
0.182
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr12_-_53448167
|
0.182
|
|
LOC283335
|
uncharacterized LOC283335
|
|
chr4_-_90758126
|
0.182
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr2_+_172950207
|
0.181
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr17_-_7197866
|
0.179
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr17_+_54671059
|
0.179
|
NM_005450
|
NOG
|
noggin
|
|
chr20_+_42143045
|
0.179
|
NM_015478
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila)
|
|
chr8_+_37887990
|
0.178
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr12_-_72666305
|
0.173
|
|
LOC283392
|
uncharacterized LOC283392
|
|
chr11_+_111783476
|
0.171
|
|
HSPB2-C11orf52
HSPB2
|
HSPB2-C11orf52 readthrough (non-protein coding)
heat shock 27kDa protein 2
|
|
chr17_+_7255427
|
0.167
|
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr1_-_150602047
|
0.167
|
|
ENSA
|
endosulfine alpha
|
|
chr9_+_100616192
|
0.166
|
|
|
|
|
chr22_-_46373007
|
0.166
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr9_-_107665959
|
0.165
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr17_-_6947241
|
0.163
|
NM_153357
|
SLC16A11
|
solute carrier family 16, member 11 (monocarboxylic acid transporter 11)
|
|
chr3_+_181430075
|
0.163
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr2_-_26205437
|
0.160
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr19_-_44285012
|
0.160
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr16_+_31044849
|
0.160
|
|
STX4
|
syntaxin 4
|
|
chr1_-_202679415
|
0.159
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr1_+_203595897
|
0.158
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr2_-_46747095
|
0.158
|
NM_080653
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2
|
|
chr14_+_29236171
|
0.157
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr19_-_1490340
|
0.156
|
|
PCSK4
|
proprotein convertase subtilisin/kexin type 4
|
|
chrX_+_38663450
|
0.156
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr5_+_173416161
|
0.155
|
NM_001144954
|
C5orf47
|
chromosome 5 open reading frame 47
|
|
chr11_-_66115016
|
0.155
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr1_-_16161279
|
0.155
|
|
|
|
|
chr1_-_55266870
|
0.153
|
NM_001114108
NM_017904
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr2_-_133427624
|
0.153
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr18_-_72920989
|
0.152
|
|
|
|
|
chr2_-_74726665
|
0.151
|
|
LBX2
|
ladybird homeobox 2
|
|
chr1_+_203596222
|
0.151
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr16_+_28889808
|
0.150
|
NM_004320
NM_173201
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr1_+_203595929
|
0.149
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chrX_-_8769377
|
0.149
|
NM_001171186
NM_174951
|
FAM9A
|
family with sequence similarity 9, member A
|
|
chr11_-_11643542
|
0.148
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr22_+_37447615
|
0.147
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr2_-_99347430
|
0.147
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr1_+_145438437
|
0.147
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr1_+_184356364
|
0.146
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr11_-_126225445
|
0.146
|
|
FLJ39051
|
uncharacterized LOC399972
|
|
chr2_+_200323088
|
0.145
|
|
|
|
|
chrX_-_3631295
|
0.145
|
|
PRKX
|
protein kinase, X-linked
|
|
chr16_+_2521499
|
0.145
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr6_-_111927082
|
0.143
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr14_+_100203998
|
0.143
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr16_-_30022300
|
0.142
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chr1_+_117452359
|
0.142
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr20_-_39766522
|
0.141
|
|
|
|
|
chr4_+_24797203
|
0.141
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr1_+_206680863
|
0.141
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr2_-_99347588
|
0.141
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chrX_+_102000906
|
0.141
|
NM_001142529
NM_001142530
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr14_-_92588065
|
0.140
|
NM_004545
|
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa
|
|
chr1_-_148176392
|
0.140
|
|
|
|
|
chr22_-_43036606
|
0.140
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr14_-_51561657
|
0.140
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr22_-_50970542
|
0.140
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr11_-_627172
|
0.140
|
NM_021920
|
SCT
|
secretin
|
|
chr8_-_41655139
|
0.139
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr3_+_39851028
|
0.139
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr3_-_46759313
|
0.139
|
NM_013270
|
PRSS50
|
protease, serine, 50
|
|
chrX_+_38663503
|
0.138
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr4_+_159091903
|
0.138
|
|
LOC285505
|
uncharacterized LOC285505
|
|
chr1_+_149804220
|
0.137
|
NM_001034077
NM_003548
|
HIST2H4B
HIST2H4A
|
histone cluster 2, H4b
histone cluster 2, H4a
|
|
chr2_-_47382425
|
0.136
|
NM_001163561
NM_173649
|
C2orf61
|
chromosome 2 open reading frame 61
|
|
chr10_-_71905947
|
0.136
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr2_+_96991934
|
0.136
|
NM_178495
NM_001163524
|
ITPRIPL1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1
|
|
chr16_+_28943259
|
0.135
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr5_-_53606303
|
0.135
|
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr6_+_167738573
|
0.134
|
NM_031949
|
TTLL2
|
tubulin tyrosine ligase-like family, member 2
|
|
chr1_-_150602089
|
0.134
|
NM_004436
NM_207042
NM_207043
NM_207044
NM_207168
|
ENSA
|
endosulfine alpha
|
|
chr6_+_1611544
|
0.133
|
|
FOXC1
|
forkhead box C1
|
|
chr6_-_3456734
|
0.132
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr20_+_62328068
|
0.132
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr3_-_12200339
|
0.131
|
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr19_+_7599590
|
0.131
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr22_+_39853534
|
0.131
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr6_+_111408780
|
0.130
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr9_-_138391696
|
0.130
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr1_-_2564428
|
0.130
|
NM_033467
|
MMEL1
|
membrane metallo-endopeptidase-like 1
|
|
chr10_+_104263726
|
0.129
|
|
SUFU
|
suppressor of fused homolog (Drosophila)
|
|
chr10_+_106028925
|
0.129
|
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr17_+_7323642
|
0.129
|
NM_199339
|
SPEM1
|
spermatid maturation 1
|
|
chr17_+_48348766
|
0.129
|
NM_001168215
|
TMEM92
|
transmembrane protein 92
|
|
chr1_-_46767367
|
0.129
|
|
|
|
|
chr2_-_219925237
|
0.129
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr11_+_65554504
|
0.128
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr16_+_67686813
|
0.128
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr7_+_139025896
|
0.128
|
NM_197964
|
C7orf55
|
chromosome 7 open reading frame 55
|
|
chr11_-_6790187
|
0.128
|
NM_001004490
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2
|
|
chr21_-_36259408
|
0.128
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr22_-_22901421
|
0.127
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr2_-_16847070
|
0.127
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr16_-_74401991
|
0.127
|
|
LOC283922
|
pyruvate dehydrogenase phosphatase regulatory subunit pseudogene
|
|
chr5_+_140855604
|
0.127
|
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr12_+_72666462
|
0.127
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr17_-_80656461
|
0.126
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr17_-_36955676
|
0.126
|
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr1_-_150602041
|
0.126
|
|
ENSA
|
endosulfine alpha
|
|
chr19_-_44285407
|
0.126
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr12_+_96588254
|
0.125
|
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr4_+_24797084
|
0.125
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr13_+_31480311
|
0.124
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr22_-_20461785
|
0.124
|
NM_015672
|
RIMBP3
|
RIMS binding protein 3
|
|
chr7_-_112726143
|
0.123
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr12_+_96588000
|
0.123
|
NM_005230
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr15_+_43809805
|
0.123
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr14_-_92587982
|
0.122
|
|
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa
|
|
chr10_-_105212022
|
0.122
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr11_-_85376061
|
0.120
|
|
CREBZF
|
CREB/ATF bZIP transcription factor
|
|
chr20_+_36149606
|
0.120
|
NM_005386
NM_181689
|
NNAT
|
neuronatin
|
|
chr2_+_18059923
|
0.119
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr12_+_109273856
|
0.118
|
NM_001917
|
DAO
|
D-amino-acid oxidase
|
|
chrX_-_20135015
|
0.118
|
NM_001168465
NM_001168466
NM_152780
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr8_-_134584012
|
0.118
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr6_-_130031278
|
0.117
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chr18_+_13218777
|
0.117
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr22_+_31892084
|
0.117
|
NM_001007467
NM_014775
|
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast)
|
|
chr18_+_44497512
|
0.117
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr17_-_76732944
|
0.115
|
|
|
|
|
chr14_-_64761077
|
0.115
|
NM_001040275
NM_001437
|
ESR2
|
estrogen receptor 2 (ER beta)
|
|
chr7_-_150946031
|
0.115
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr9_-_98279246
|
0.114
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr11_-_67407024
|
0.114
|
NM_005995
|
TBX10
|
T-box 10
|
|
chr22_+_38201300
|
0.114
|
|
H1F0
|
H1 histone family, member 0
|
|
chr6_-_168479796
|
0.113
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr10_+_104263716
|
0.112
|
NM_001178133
NM_016169
|
SUFU
|
suppressor of fused homolog (Drosophila)
|
|
chr3_+_6902812
|
0.112
|
|
GRM7
|
glutamate receptor, metabotropic 7
|
|
chr2_-_16081703
|
0.112
|
|
MYCNOS
|
MYCN opposite strand/antisense RNA (non-protein coding)
|
|
chr12_+_112451229
|
0.112
|
|
|
|
|
chr4_-_159092509
|
0.111
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr22_+_31892372
|
0.111
|
|
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast)
|
|
chr5_+_145718586
|
0.111
|
NM_002700
|
POU4F3
|
POU class 4 homeobox 3
|
|
chr19_-_46405854
|
0.110
|
NM_001012643
|
MYPOP
|
Myb-related transcription factor, partner of profilin
|
|
chr1_-_43205733
|
0.110
|
NM_001123395
NM_001185117
NM_148960
|
CLDN19
|
claudin 19
|
|
chr12_-_103310896
|
0.109
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr19_-_50354906
|
0.109
|
|
LOC100506033
|
uncharacterized LOC100506033
|
|
chr6_-_134373618
|
0.109
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr8_+_42010463
|
0.109
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr20_+_32319765
|
0.109
|
NM_032819
|
ZNF341
|
zinc finger protein 341
|
|
chr19_-_12886421
|
0.108
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr22_+_50919970
|
0.107
|
NM_024866
NM_001253845
|
ADM2
|
adrenomedullin 2
|
|
chr3_+_113616317
|
0.107
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr14_-_50778788
|
0.107
|
NM_024884
|
L2HGDH
|
L-2-hydroxyglutarate dehydrogenase
|
|
chr1_-_41328002
|
0.107
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|