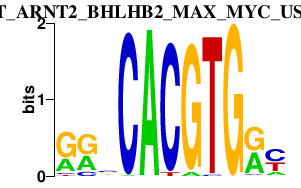
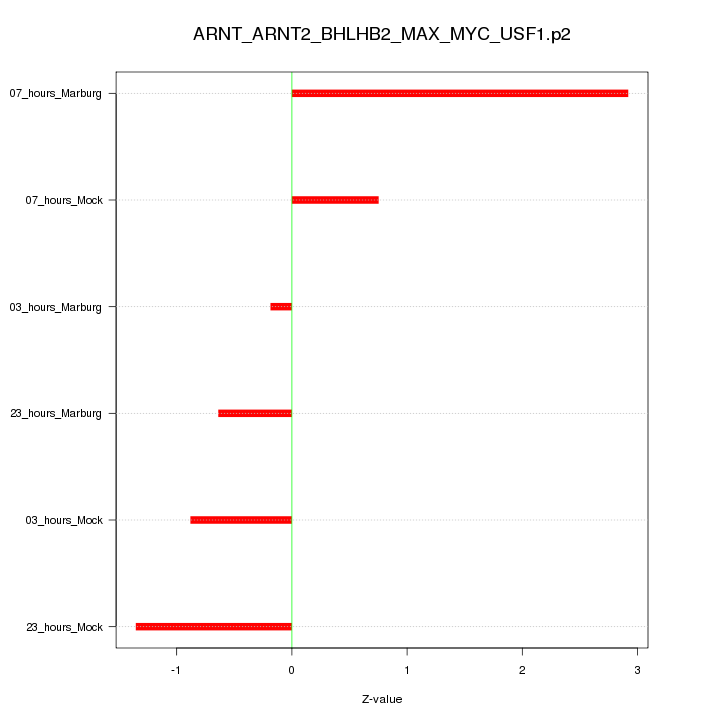
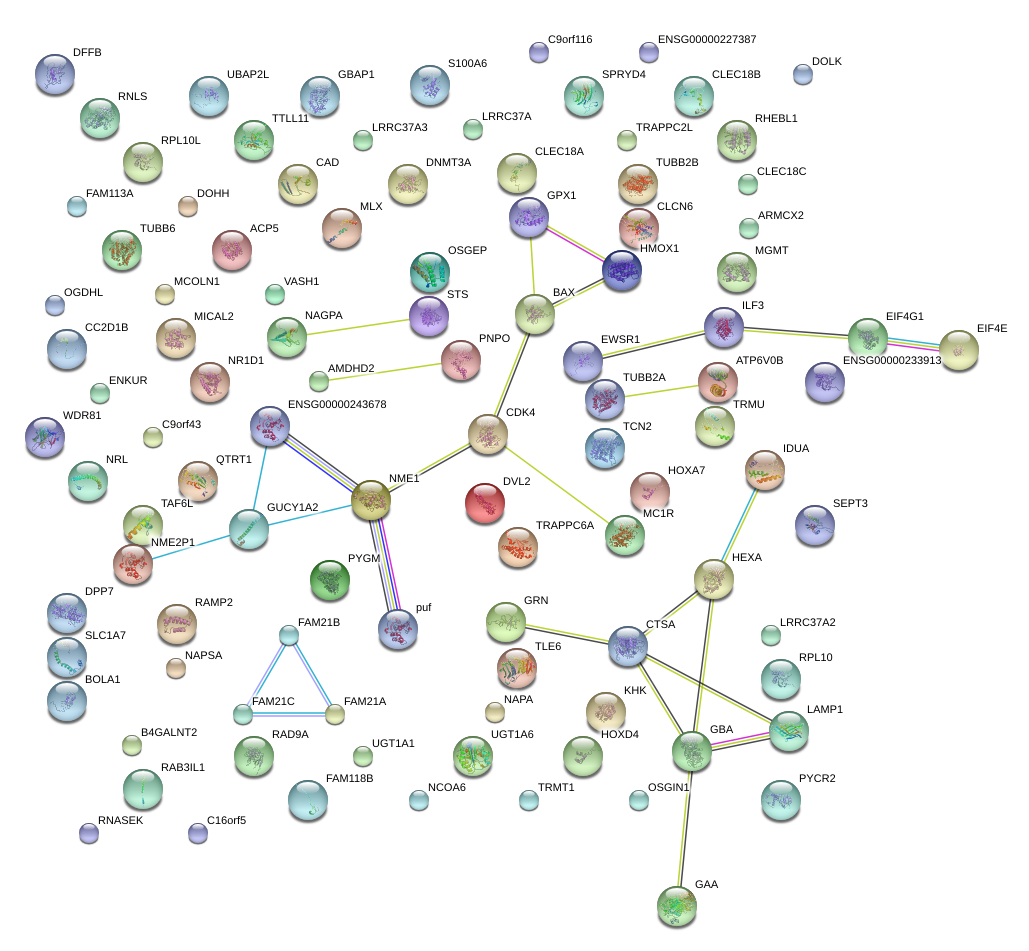
|
chr17_-_7137722
|
0.839
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr12_+_112451229
|
0.825
|
|
|
|
|
chr14_-_47120940
|
0.810
|
NM_080746
|
RPL10L
|
ribosomal protein L10-like
|
|
chr16_+_2570322
|
0.797
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr3_-_40350948
|
0.749
|
|
FLJ33065
|
uncharacterized LOC440952
|
|
chr20_+_44520021
|
0.718
|
|
CTSA
|
cathepsin A
|
|
chr1_+_154193294
|
0.700
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr20_+_44519965
|
0.690
|
|
CTSA
|
cathepsin A
|
|
chr9_-_138391696
|
0.677
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr17_-_7137580
|
0.675
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr6_-_13615386
|
0.624
|
|
|
|
|
chr22_+_35777087
|
0.614
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr17_+_79885567
|
0.613
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr2_-_200715895
|
0.596
|
|
FONG
|
uncharacterized LOC348751
|
|
chr17_+_6915952
|
0.592
|
|
RNASEK
|
ribonuclease, RNase K
|
|
chr10_+_99344421
|
0.588
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr11_+_67159421
|
0.586
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr20_+_44519924
|
0.583
|
|
CTSA
|
cathepsin A
|
|
chr19_+_49458151
|
0.581
|
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_226111977
|
0.563
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr7_-_27196163
|
0.557
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr2_+_177016112
|
0.553
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr2_+_234668914
|
0.552
|
NM_000463
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr17_+_78075360
|
0.547
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr19_+_2977480
|
0.535
|
NM_001143986
NM_024760
|
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila)
|
|
chr19_+_10812111
|
0.534
|
NM_031209
|
QTRT1
|
queuine tRNA-ribosyltransferase 1
|
|
chr19_+_49458031
|
0.528
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr9_-_140009167
|
0.526
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr1_-_155210887
|
0.524
|
|
GBA
|
glucosidase, beta, acid
|
|
chr10_+_51827710
|
0.512
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr11_-_61684981
|
0.494
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr17_+_78075390
|
0.493
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr1_-_155211049
|
0.486
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr2_+_27440307
|
0.482
|
|
|
|
|
chr17_+_1628032
|
0.469
|
NM_001163809
|
WDR81
|
WD repeat domain 81
|
|
chr19_+_49458129
|
0.464
|
|
BAX
|
BCL2-associated X protein
|
|
chr9_-_140008956
|
0.458
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr12_-_49463729
|
0.457
|
NM_144593
|
RHEBL1
|
Ras homolog enriched in brain like 1
|
|
chr17_+_78075299
|
0.452
|
NM_000152
NM_001079803
NM_001079804
|
GAA
|
glucosidase, alpha; acid
|
|
chr17_+_6915735
|
0.446
|
NM_001004333
|
RNASEK
|
ribonuclease, RNase K
|
|
chr12_-_58146118
|
0.442
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr19_-_3500620
|
0.439
|
NM_031304
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase
|
|
chr20_+_44520203
|
0.438
|
|
CTSA
|
cathepsin A
|
|
chr12_-_58146110
|
0.438
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr16_+_83986826
|
0.435
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr11_+_62538774
|
0.424
|
NM_006473
|
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr2_+_177016337
|
0.422
|
|
|
|
|
chr17_+_42422490
|
0.418
|
NM_002087
|
GRN
|
granulin
|
|
chr9_+_116172934
|
0.413
|
NM_152786
|
C9orf43
|
chromosome 9 open reading frame 43
|
|
chr2_+_27440342
|
0.403
|
|
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
|
|
chr9_-_131709783
|
0.402
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr10_-_25304865
|
0.401
|
NM_145010
|
ENKUR
|
enkurin, TRPC channel interacting protein
|
|
chr3_+_194207685
|
0.400
|
|
FLJ34208
|
uncharacterized LOC401106
|
|
chr17_-_62915580
|
0.399
|
|
LRRC37A3
|
leucine rich repeat containing 37, member A3
|
|
chr19_-_45681454
|
0.399
|
NM_024108
|
TRAPPC6A
|
trafficking protein particle complex 6A
|
|
chr22_+_35777051
|
0.393
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr10_+_51827654
|
0.391
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr14_-_24584125
|
0.391
|
|
NRL
|
neural retina leucine zipper
|
|
chr12_-_58146108
|
0.389
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_+_28343390
|
0.388
|
|
|
|
|
chr16_-_4588738
|
0.387
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr1_-_53608120
|
0.385
|
NM_006671
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chr11_-_106888829
|
0.383
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr11_-_71814268
|
0.382
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr3_-_49395697
|
0.382
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr12_-_58146025
|
0.378
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr20_-_2821245
|
0.377
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr1_-_153508597
|
0.377
|
NM_014624
|
S100A6
|
S100 calcium binding protein A6
|
|
chr12_+_90102639
|
0.369
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chr2_+_27309610
|
0.369
|
NM_000221
NM_006488
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr14_-_20923266
|
0.368
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr16_-_74455239
|
0.365
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr11_+_126081618
|
0.365
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr4_+_980775
|
0.362
|
NM_000203
|
IDUA
|
iduronidase, alpha-L-
|
|
chr17_-_7137675
|
0.355
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_-_13227212
|
0.355
|
NM_001142554
NM_017722
NM_001136035
|
TRMT1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr1_+_11866302
|
0.353
|
|
CLCN6
|
chloride channel 6
|
|
chr20_-_2821120
|
0.349
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr19_-_11688508
|
0.347
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr16_+_88923490
|
0.345
|
NM_016209
|
TRAPPC2L
|
trafficking protein particle complex 2-like
|
|
chr13_+_113951537
|
0.345
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr17_+_40913211
|
0.343
|
NM_005854
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr10_-_50970321
|
0.343
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr13_+_113951589
|
0.342
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr17_-_38256548
|
0.340
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr17_+_46019022
|
0.338
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr10_+_131265453
|
0.335
|
NM_002412
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr3_+_184032414
|
0.335
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr11_+_12132063
|
0.334
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr20_+_44519590
|
0.332
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr15_-_72668301
|
0.330
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr1_+_149871154
|
0.327
|
NM_016074
|
BOLA1
|
bolA homolog 1 (E. coli)
|
|
chr1_-_52831817
|
0.327
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr22_+_29663997
|
0.322
|
NM_001163285
NM_001163286
NM_001163287
NM_005243
NM_013986
|
EWSR1
|
Ewing sarcoma breakpoint region 1
|
|
chr19_+_7587436
|
0.321
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chr3_+_184032319
|
0.321
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_+_56862300
|
0.320
|
NM_207344
|
SPRYD4
|
SPRY domain containing 4
|
|
chr17_+_40719064
|
0.319
|
NM_170607
NM_198204
NM_198205
|
MLX
|
MAX-like protein X
|
|
chr2_-_25474948
|
0.318
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chrX_-_100914816
|
0.318
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr19_-_48018277
|
0.313
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr19_+_10765106
|
0.311
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr2_+_172864803
|
0.309
|
NM_199227
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial)
|
|
chr16_-_5083933
|
0.308
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr1_+_44440595
|
0.307
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr16_+_89985268
|
0.304
|
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor)
|
|
chrX_+_7065820
|
0.301
|
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr17_+_42422627
|
0.301
|
|
GRN
|
granulin
|
|
chr19_+_10764987
|
0.300
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr3_-_49395751
|
0.298
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr22_+_31003069
|
0.298
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr14_+_77228940
|
0.297
|
|
VASH1
|
vasohibin 1
|
|
chr10_-_90342947
|
0.296
|
NM_001031709
NM_018363
|
RNLS
|
renalase, FAD-dependent amine oxidase
|
|
chr10_+_131265500
|
0.292
|
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr1_+_11866206
|
0.289
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr9_-_124855559
|
0.288
|
|
TTLL11
|
tubulin tyrosine ligase-like family, member 11
|
|
chr4_-_99850242
|
0.286
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr22_+_42372763
|
0.285
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr17_+_49244082
|
0.285
|
NM_001018138
|
NME2
|
non-metastatic cells 2, protein (NM23B) expressed in
|
|
chr8_+_104427590
|
0.284
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chr3_+_127317274
|
0.283
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr17_+_46970128
|
0.283
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr14_-_75389144
|
0.281
|
NM_031464
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1
|
|
chr3_-_51975879
|
0.281
|
NM_004704
|
RRP9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast)
|
|
chr6_+_37137882
|
0.281
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr1_-_28559513
|
0.280
|
NM_014280
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr22_-_39268230
|
0.278
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr19_+_42364262
|
0.274
|
|
RPS19
|
ribosomal protein S19
|
|
chr12_+_117176085
|
0.273
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr11_+_113659924
|
0.272
|
|
LOC390251
|
SH3-domain GRB2-like 1 pseudogene
|
|
chr19_-_51307865
|
0.272
|
NM_199250
NM_199249
|
C19orf48
|
chromosome 19 open reading frame 48
|
|
chr3_+_127317271
|
0.271
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr1_-_154193073
|
0.270
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr22_-_20104745
|
0.268
|
NM_022727
NM_182984
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr17_+_35306174
|
0.267
|
NM_012138
|
AATF
|
apoptosis antagonizing transcription factor
|
|
chr19_+_42364322
|
0.267
|
|
RPS19
|
ribosomal protein S19
|
|
chr19_+_10764896
|
0.265
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr9_+_131709970
|
0.265
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr3_+_127317212
|
0.262
|
NM_004526
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr12_+_56109851
|
0.261
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr16_-_4588379
|
0.259
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr21_+_45209398
|
0.258
|
NM_003683
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr1_-_1710228
|
0.257
|
NM_001198993
|
NADK
|
NAD kinase
|
|
chr11_-_119252364
|
0.257
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_-_71221833
|
0.254
|
|
TEX261
|
testis expressed 261
|
|
chr3_-_187463474
|
0.254
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr8_+_42128834
|
0.253
|
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr16_+_699277
|
0.253
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr9_+_131709991
|
0.253
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr16_-_1524926
|
0.252
|
|
CLCN7
|
chloride channel 7
|
|
chr9_+_103115342
|
0.252
|
|
|
|
|
chr20_-_2821789
|
0.251
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr19_-_11688484
|
0.251
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_-_12886263
|
0.250
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chrX_+_152224862
|
0.249
|
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr21_+_45209448
|
0.249
|
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr3_-_187463227
|
0.249
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr12_+_112451261
|
0.248
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr2_-_241497413
|
0.246
|
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|
|
chr11_-_119252267
|
0.246
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_+_27440233
|
0.245
|
NM_004341
|
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
|
|
chr1_+_44440623
|
0.244
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr16_+_84002099
|
0.242
|
NM_019065
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr8_-_38034168
|
0.241
|
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr19_-_48018163
|
0.241
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr17_+_42422665
|
0.241
|
|
GRN
|
granulin
|
|
chr1_+_45792544
|
0.241
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr22_-_29663823
|
0.240
|
NM_012265
|
RHBDD3
|
rhomboid domain containing 3
|
|
chr4_-_100242487
|
0.240
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr17_+_40688264
|
0.239
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr1_-_27226928
|
0.238
|
|
GPATCH3
|
G patch domain containing 3
|
|
chr19_+_56166418
|
0.238
|
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chr1_-_11865963
|
0.237
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr16_+_22308722
|
0.235
|
NM_018119
|
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD)
|
|
chr11_-_64014407
|
0.235
|
NM_138689
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chr11_-_66335958
|
0.233
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr1_-_110950241
|
0.233
|
|
HBXIP
|
hepatitis B virus x interacting protein
|
|
chr1_+_11866240
|
0.232
|
|
CLCN6
|
chloride channel 6
|
|
chr1_-_27226937
|
0.231
|
NM_022078
|
GPATCH3
|
G patch domain containing 3
|
|
chr5_+_154134853
|
0.231
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr1_-_204329043
|
0.231
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr3_+_33318911
|
0.228
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr1_+_44440673
|
0.228
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr2_-_240323332
|
0.227
|
|
HDAC4
|
histone deacetylase 4
|
|
chr1_+_154193757
|
0.227
|
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr17_+_48172508
|
0.227
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr13_+_113951461
|
0.225
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr17_+_73975297
|
0.225
|
NM_001113324
|
TEN1
TEN1-CDK3
|
TEN1 telomerase capping complex subunit homolog (S. cerevisiae)
TEN1-CDK3 readthrough
|
|
chr16_+_89778242
|
0.225
|
|
LOC100128881
|
uncharacterized LOC100128881
|
|
chr19_-_49140569
|
0.224
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr6_+_42018264
|
0.222
|
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr1_+_180124038
|
0.221
|
|
|
|
|
chr2_-_241497404
|
0.219
|
NM_016552
NM_017844
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|
|
chr1_+_226016428
|
0.219
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr16_-_5083913
|
0.218
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr3_+_23986735
|
0.217
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr19_-_45909566
|
0.217
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr9_-_124855884
|
0.217
|
NM_001139442
NM_194252
|
TTLL11
|
tubulin tyrosine ligase-like family, member 11
|
|
chr2_+_223916861
|
0.216
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr17_-_48450532
|
0.216
|
NM_016504
|
MRPL27
|
mitochondrial ribosomal protein L27
|