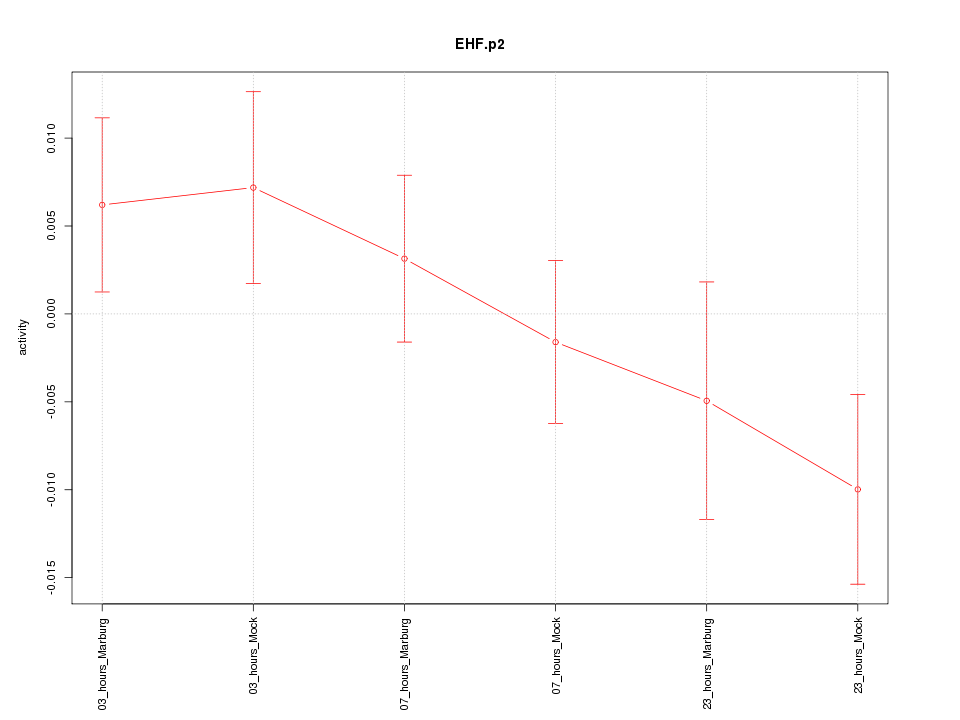
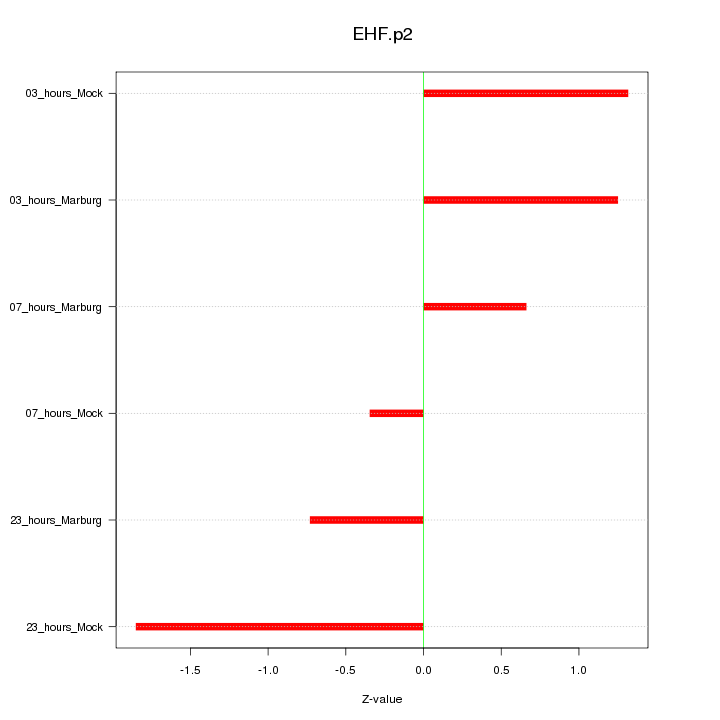
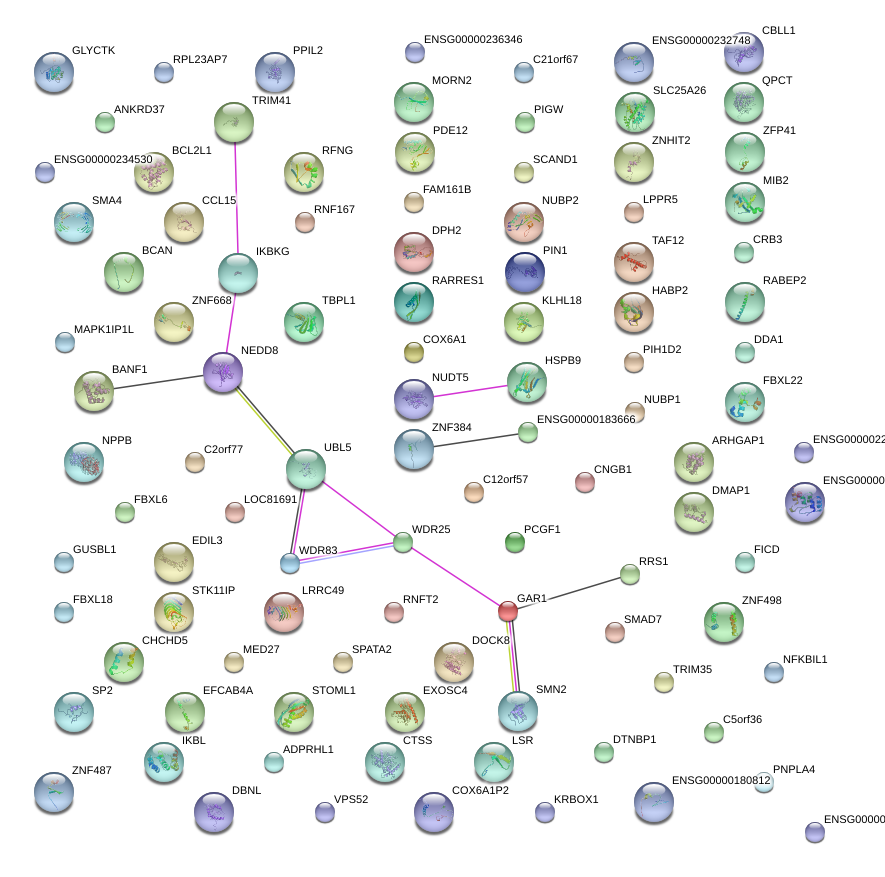
|
chr6_+_31515352
|
0.645
|
NM_001144961
NM_005007
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chr2_+_39103102
|
0.582
|
NM_001145450
|
MORN2
|
MORN repeat containing 2
|
|
chr6_-_15663098
|
0.574
|
|
DTNBP1
|
dystrobrevin binding protein 1
|
|
chr5_+_69696805
|
0.563
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr8_+_144349564
|
0.527
|
NM_138465
|
GLI4
|
GLI family zinc finger 4
|
|
chr11_+_65769886
|
0.511
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr3_-_20053618
|
0.511
|
NM_001252657
|
PP2D1
|
protein phosphatase 2C-like domain containing 1
|
|
chr11_-_46722115
|
0.485
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr9_+_214816
|
0.468
|
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr3_+_57542054
|
0.463
|
|
PDE12
|
phosphodiesterase 12
|
|
chr11_-_46722139
|
0.462
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chrX_-_7895424
|
0.459
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr2_+_113341999
|
0.445
|
NM_032309
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr20_-_34542205
|
0.445
|
|
SCAND1
|
SCAN domain containing 1
|
|
chr3_-_169482847
|
0.441
|
|
TERC
|
telomerase RNA component
|
|
chr21_-_46359685
|
0.438
|
|
C21orf67
|
chromosome 21 open reading frame 67
|
|
chr11_-_46722160
|
0.432
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr17_+_45973515
|
0.419
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr3_+_42977833
|
0.409
|
NM_001205272
|
KRBOX1
|
KRAB box domain containing 1
|
|
chr1_-_99470177
|
0.408
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr2_+_113342171
|
0.398
|
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chrX_-_7895764
|
0.396
|
NM_001142389
NM_001172672
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr15_+_71184781
|
0.391
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr11_+_65769956
|
0.383
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr1_-_150738255
|
0.382
|
|
CTSS
|
cathepsin S
|
|
chr2_-_114384630
|
0.371
|
|
RPL23AP7
|
ribosomal protein L23a pseudogene 7
|
|
chr1_-_1551041
|
0.360
|
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr5_+_180650274
|
0.359
|
|
TRIM41
|
tripartite motif containing 41
|
|
chr6_-_13615386
|
0.357
|
|
|
|
|
chr6_+_134274247
|
0.348
|
NM_004865
|
TBPL1
|
TBP-like 1
|
|
chr12_-_6798619
|
0.341
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr11_-_64885124
|
0.337
|
NM_014205
|
ZNHIT2
|
zinc finger, HIT-type containing 2
|
|
chr16_+_20817766
|
0.335
|
NM_001144924
NM_001199053
NM_030941
|
LOC81691
|
exonuclease NEF-sp
|
|
chr11_-_111944757
|
0.330
|
NM_001082619
NM_138789
|
PIH1D2
|
PIH1 domain containing 2
|
|
chr17_+_34891340
|
0.325
|
NM_178517
|
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W
|
|
chr2_-_170550859
|
0.323
|
NM_001085447
|
C2orf77
|
chromosome 2 open reading frame 77
|
|
chr10_-_12238053
|
0.321
|
NM_014142
|
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5
|
|
chr14_-_24701472
|
0.320
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr12_-_123201319
|
0.319
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr12_-_6798528
|
0.318
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr8_-_146052424
|
0.318
|
|
LOC100130027
|
uncharacterized LOC100130027
|
|
chr16_-_28936454
|
0.315
|
NM_024816
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2
|
|
chr19_+_12780516
|
0.309
|
NM_032332
|
WDR83
|
WD repeat domain 83
|
|
chr10_+_43932573
|
0.307
|
|
ZNF487P
|
zinc finger protein 487, pseudogene
|
|
chr20_-_48532067
|
0.304
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr5_+_180650233
|
0.303
|
NM_033549
NM_201627
|
TRIM41
|
tripartite motif containing 41
|
|
chr19_+_6464259
|
0.302
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr8_-_27168783
|
0.295
|
NM_171982
|
TRIM35
|
tripartite motif containing 35
|
|
chr17_-_80009649
|
0.291
|
NM_002917
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr16_+_20818010
|
0.290
|
|
LOC81691
|
exonuclease NEF-sp
|
|
chr7_+_44084317
|
0.290
|
|
DBNL
|
drebrin-like
|
|
chr4_-_103422436
|
0.289
|
|
|
|
|
chr2_-_70314065
|
0.289
|
|
PCBP1-AS1
|
PCBP1 antisense RNA 1 (non-protein coding)
|
|
chr12_-_6798419
|
0.288
|
|
ZNF384
|
zinc finger protein 384
|
|
chr12_+_108909049
|
0.286
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr3_+_66271167
|
0.285
|
NM_001164796
|
SLC25A26
|
solute carrier family 25, member 26
|
|
chr3_+_52321835
|
0.284
|
NM_001144951
NM_145262
|
GLYCTK
|
glycerate kinase
|
|
chr8_+_67341244
|
0.282
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr17_+_4843883
|
0.281
|
|
RNF167
|
ring finger protein 167
|
|
chr3_+_57541973
|
0.281
|
NM_177966
|
PDE12
|
phosphodiesterase 12
|
|
chr1_-_28969516
|
0.280
|
NM_001135218
NM_005644
|
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa
|
|
chr18_-_46475702
|
0.280
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr16_-_31085073
|
0.277
|
|
ZNF668
|
zinc finger protein 668
|
|
chr8_+_145133567
|
0.276
|
|
EXOSC4
|
exosome component 4
|
|
chr3_+_52321853
|
0.275
|
|
GLYCTK
|
glycerate kinase
|
|
chr11_+_65769773
|
0.271
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr14_+_100842728
|
0.269
|
NM_001161476
NM_024515
|
WDR25
|
WD repeat domain 25
|
|
chr7_+_99214553
|
0.268
|
NM_145115
|
ZNF498
|
zinc finger protein 498
|
|
chr14_-_24701561
|
0.267
|
NM_001199823
NM_006156
|
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr1_-_32687945
|
0.267
|
|
TMEM234
|
transmembrane protein 234
|
|
chr14_+_35343401
|
0.265
|
|
|
|
|
chr7_+_107384573
|
0.263
|
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr6_-_15663260
|
0.261
|
NM_032122
NM_183040
|
DTNBP1
|
dystrobrevin binding protein 1
|
|
chr2_+_37571752
|
0.261
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr11_+_1331048
|
0.259
|
|
LOC255512
|
uncharacterized LOC255512
|
|
chr2_-_27616443
|
0.258
|
|
FTH1P3
|
ferritin, heavy polypeptide 1 pseudogene 3
|
|
chr16_+_2653350
|
0.252
|
|
LOC652276
|
potassium channel tetramerisation domain containing 5 pseudogene
|
|
chr16_+_10837673
|
0.249
|
NM_002484
|
NUBP1
|
nucleotide binding protein 1
|
|
chr13_-_114103452
|
0.248
|
NM_199162
|
ADPRHL1
|
ADP-ribosylhydrolase like 1
|
|
chr21_+_46359966
|
0.248
|
|
|
|
|
chr1_+_44679111
|
0.247
|
NM_001034023
NM_001034024
NM_019100
|
DMAP1
|
DNA methyltransferase 1 associated protein 1
|
|
chr5_+_180650709
|
0.246
|
|
TRIM41
|
tripartite motif containing 41
|
|
chrX_+_153775561
|
0.244
|
NM_001099857
NM_001145255
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr19_+_9938587
|
0.244
|
|
UBL5
|
ubiquitin-like 5
|
|
chr4_+_186317964
|
0.243
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr15_+_63889551
|
0.242
|
NM_203373
|
FBXL22
|
F-box and leucine-rich repeat protein 22
|
|
chr20_-_48532036
|
0.240
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr4_+_110736911
|
0.240
|
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr7_-_5553305
|
0.239
|
NM_024963
|
FBXL18
|
F-box and leucine-rich repeat protein 18
|
|
chr10_+_115310589
|
0.239
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr17_+_40274755
|
0.237
|
NM_033194
|
HSPB9
|
heat shock protein, alpha-crystallin-related, B9
|
|
chr20_-_48531986
|
0.237
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr19_+_17420352
|
0.235
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr17_-_34328530
|
0.235
|
|
CCL14-CCL15
CCL15
|
CCL14-CCL15 readthrough
chemokine (C-C motif) ligand 15
|
|
chr14_+_55518313
|
0.233
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr9_-_98638243
|
0.233
|
|
LINC00476
|
long intergenic non-protein coding RNA 476
|
|
chr10_-_12237840
|
0.233
|
|
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5
|
|
chr19_+_35739838
|
0.232
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr3_-_158450205
|
0.232
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr2_+_220462560
|
0.231
|
NM_052902
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr15_-_74284518
|
0.230
|
NM_004809
|
STOML1
|
stomatin (EPB72)-like 1
|
|
chr6_-_33239585
|
0.230
|
NM_022553
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae)
|
|
chr5_-_83680191
|
0.229
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr17_+_56065408
|
0.229
|
|
|
|
|
chr12_+_7053180
|
0.228
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr2_-_74734498
|
0.227
|
|
PCGF1
|
polycomb group ring finger 1
|
|
chr20_-_30310597
|
0.225
|
NM_001191
NM_138578
|
BCL2L1
|
BCL2-like 1
|
|
chr11_-_65337743
|
0.224
|
|
LOC254100
|
uncharacterized LOC254100
|
|
chr11_+_827584
|
0.222
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr12_+_117176085
|
0.222
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr5_-_93954176
|
0.221
|
NM_001145678
NM_173665
|
KIAA0825
|
KIAA0825
|
|
chr9_-_134955056
|
0.220
|
|
MED27
|
mediator complex subunit 27
|
|
chr3_+_47324329
|
0.220
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr22_+_22020272
|
0.219
|
NM_014337
NM_148175
NM_148176
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr1_+_44435649
|
0.216
|
NM_001039589
NM_001384
|
DPH2
|
DPH2 homolog (S. cerevisiae)
|
|
chr14_-_74417116
|
0.215
|
NM_152445
|
FAM161B
|
family with sequence similarity 161, member B
|
|
chr14_-_74416711
|
0.214
|
|
FAM161B
|
family with sequence similarity 161, member B
|
|
chr18_-_72264808
|
0.212
|
|
LOC400657
|
uncharacterized LOC400657
|
|
chr14_-_24701559
|
0.211
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr19_-_12780206
|
0.210
|
|
WDR83OS
|
WD repeat domain 83 opposite strand
|
|
chr1_-_11918889
|
0.209
|
NM_002521
|
NPPB
|
natriuretic peptide B
|
|
chr14_+_55518398
|
0.209
|
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr12_+_120875891
|
0.209
|
NM_004373
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1
|
|
chr12_-_56360457
|
0.209
|
NM_006928
|
PMEL
|
premelanosome protein
|
|
chr15_+_74753602
|
0.208
|
|
LOC440288
|
uncharacterized LOC440288
|
|
chr5_-_69881965
|
0.208
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr6_+_30525050
|
0.206
|
|
PRR3
|
proline rich 3
|
|
chr17_-_33288475
|
0.205
|
NM_001193529
NM_001193530
NM_006584
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2)
|
|
chr5_-_40835325
|
0.205
|
NM_000997
|
RPL37
|
ribosomal protein L37
|
|
chr18_-_72265038
|
0.205
|
|
LOC400657
|
uncharacterized LOC400657
|
|
chr1_+_95285900
|
0.204
|
NM_001114106
|
SLC44A3
|
solute carrier family 44, member 3
|
|
chrX_-_153775774
|
0.204
|
NM_001042351
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr9_-_100684770
|
0.203
|
NM_016481
|
C9orf156
|
chromosome 9 open reading frame 156
|
|
chr6_-_32143899
|
0.203
|
NM_006411
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr17_+_34890796
|
0.202
|
|
|
|
|
chr14_+_93673622
|
0.202
|
|
UBR7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative)
|
|
chr1_-_169336999
|
0.200
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr16_+_3493605
|
0.200
|
NM_001083601
|
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit
|
|
chr11_+_576482
|
0.200
|
NM_020901
|
PHRF1
|
PHD and ring finger domains 1
|
|
chr19_-_56632644
|
0.200
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr3_-_130745583
|
0.200
|
NM_014065
|
ASTE1
|
asteroid homolog 1 (Drosophila)
|
|
chr6_-_32143795
|
0.199
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr19_+_56152281
|
0.199
|
NM_207115
|
ZNF580
|
zinc finger protein 580
|
|
chr14_+_35761543
|
0.198
|
NM_002791
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6
|
|
chr17_+_260092
|
0.198
|
NM_001013672
|
C17orf97
|
chromosome 17 open reading frame 97
|
|
chr17_-_4843204
|
0.197
|
NM_001165417
NM_001165418
NM_003562
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11
|
|
chr20_+_42984088
|
0.197
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr11_-_64764388
|
0.197
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chr9_-_127703294
|
0.196
|
NM_002077
|
GOLGA1
|
golgin A1
|
|
chr9_-_134955243
|
0.196
|
NM_004269
|
CRSP8P
MED27
|
mediator complex subunit 27 pseudogene
mediator complex subunit 27
|
|
chr14_+_55518372
|
0.194
|
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr10_+_129705359
|
0.194
|
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr12_+_51632654
|
0.193
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr6_+_28092386
|
0.193
|
NM_025231
|
ZSCAN16
|
zinc finger and SCAN domain containing 16
|
|
chr1_+_762987
|
0.192
|
|
LOC643837
|
uncharacterized LOC643837
|
|
chr17_-_11900688
|
0.191
|
NM_144680
|
ZNF18
|
zinc finger protein 18
|
|
chr1_-_92949045
|
0.190
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr19_-_12780448
|
0.190
|
NM_016145
|
WDR83OS
|
WD repeat domain 83 opposite strand
|
|
chr6_-_32143886
|
0.190
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chrX_+_148622518
|
0.189
|
NM_001171907
NM_001171908
NM_178124
|
CXorf40A
|
chromosome X open reading frame 40A
|
|
chr14_+_67827060
|
0.189
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr17_-_58156274
|
0.189
|
NM_022070
|
HEATR6
|
HEAT repeat containing 6
|
|
chr19_+_56166418
|
0.189
|
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chr15_+_90895476
|
0.187
|
NM_001004309
|
ZNF774
|
zinc finger protein 774
|
|
chrX_-_154250769
|
0.187
|
NM_000132
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr2_+_128458943
|
0.187
|
|
SFT2D3
|
SFT2 domain containing 3
|
|
chr15_-_75135524
|
0.187
|
NM_001099436
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr14_+_45431333
|
0.187
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr19_+_18682613
|
0.186
|
NM_001033930
NM_003333
|
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1
|
|
chr6_-_33239439
|
0.185
|
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae)
|
|
chr7_-_72722786
|
0.185
|
NM_001168347
NM_001168348
NM_018044
NM_148956
|
NSUN5
|
NOP2/Sun domain family, member 5
|
|
chr19_+_41816032
|
0.185
|
NM_052848
|
CCDC97
|
coiled-coil domain containing 97
|
|
chr16_+_89724175
|
0.185
|
NM_153025
|
C16orf55
|
chromosome 16 open reading frame 55
|
|
chr7_-_73097729
|
0.184
|
NM_032317
|
DNAJC30
|
DnaJ (Hsp40) homolog, subfamily C, member 30
|
|
chr7_+_74306895
|
0.184
|
|
PMS2P5
|
postmeiotic segregation increased 2 pseudogene 5
|
|
chr1_-_153508597
|
0.184
|
NM_014624
|
S100A6
|
S100 calcium binding protein A6
|
|
chr3_-_151987332
|
0.183
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr8_-_27168753
|
0.183
|
|
TRIM35
|
tripartite motif containing 35
|
|
chr21_+_47706297
|
0.183
|
|
YBEY
|
ybeY metallopeptidase (putative)
|
|
chr21_-_37432789
|
0.183
|
NM_017438
|
SETD4
|
SET domain containing 4
|
|
chr6_+_33239855
|
0.182
|
|
RPS18
|
ribosomal protein S18
|
|
chr5_-_40835283
|
0.182
|
|
RPL37
SNORD72
|
ribosomal protein L37
small nucleolar RNA, C/D box 72
|
|
chr9_+_136223414
|
0.182
|
NM_017503
|
SURF2
|
surfeit 2
|
|
chr22_-_24092951
|
0.182
|
NM_021916
|
ZNF70
|
zinc finger protein 70
|
|
chr4_+_668250
|
0.181
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr4_-_171011292
|
0.181
|
NM_182662
|
AADAT
|
aminoadipate aminotransferase
|
|
chr11_+_71791376
|
0.181
|
NM_001145307
NM_001145308
NM_001205138
NM_145309
|
LRTOMT
|
leucine rich transmembrane and 0-methyltransferase domain containing
|
|
chr17_-_3571819
|
0.181
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr17_+_4843585
|
0.181
|
NM_015528
|
RNF167
|
ring finger protein 167
|
|
chr5_+_157158322
|
0.180
|
NM_017872
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae)
|
|
chr21_+_43933983
|
0.180
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr19_-_11616395
|
0.180
|
|
ZNF653
|
zinc finger protein 653
|
|
chr17_-_42264023
|
0.179
|
|
C17orf65
|
chromosome 17 open reading frame 65
|
|
chr14_+_75179846
|
0.179
|
NM_015962
|
FCF1
|
FCF1 small subunit (SSU) processome component homolog (S. cerevisiae)
|
|
chr2_-_241500459
|
0.179
|
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|
|
chr17_-_33288415
|
0.178
|
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2)
|
|
chr2_+_8822112
|
0.178
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr12_+_53845887
|
0.178
|
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr1_+_22379119
|
0.178
|
NM_001039802
NM_001791
NM_044472
|
CDC42
|
cell division cycle 42 (GTP binding protein, 25kDa)
|
|
chr2_-_69870837
|
0.177
|
|
AAK1
|
AP2 associated kinase 1
|