|
chr1_-_183559463
|
2.040
|
|
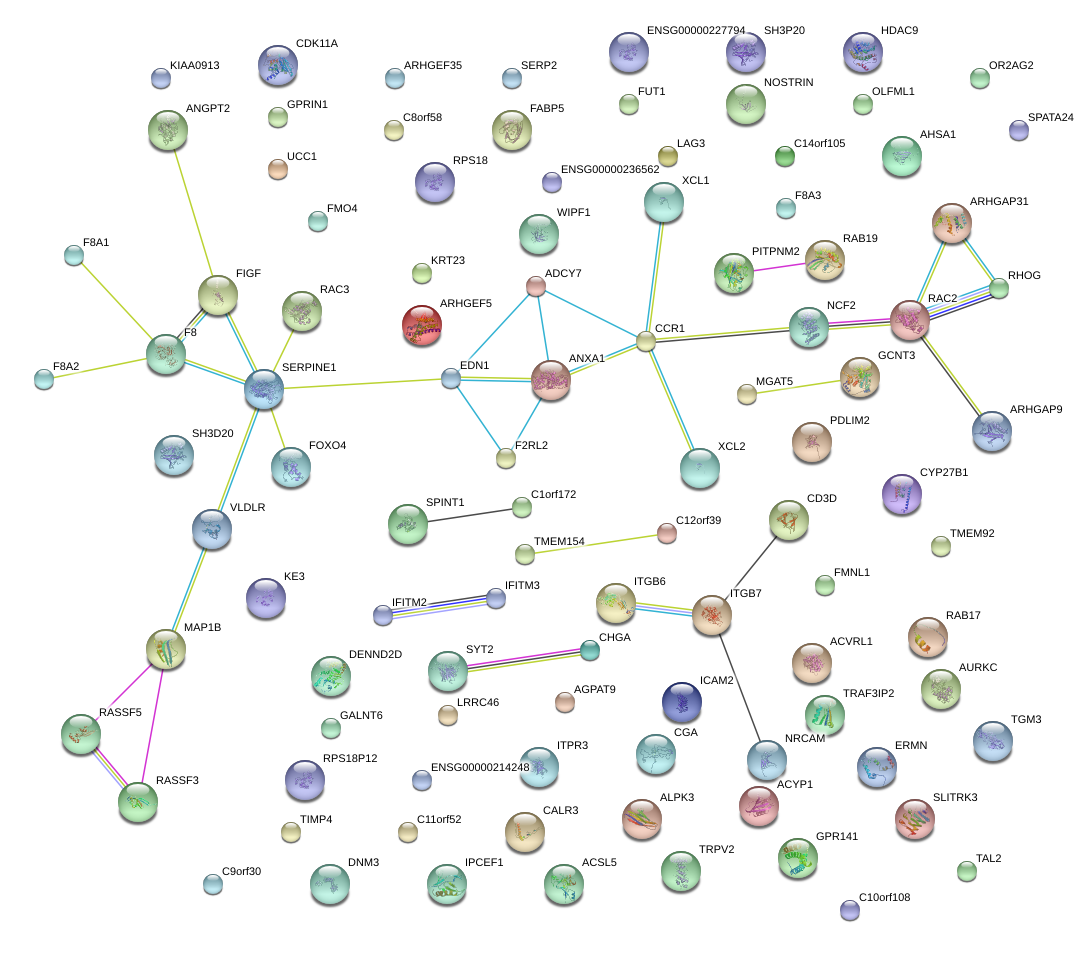
NCF2
|
neutrophil cytosolic factor 2
|
|
chr1_-_183559669
|
1.694
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr5_+_87564840
|
1.304
|
|
LOC100505894
|
uncharacterized LOC100505894
|
|
chr1_-_1590403
|
1.253
|
|
CDK11A
CDK11B
|
cyclin-dependent kinase 11A
cyclin-dependent kinase 11B
|
|
chr1_-_183560010
|
1.198
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr5_-_138739755
|
1.050
|
NM_194296
|
SPATA24
|
spermatogenesis associated 24
|
|
chr2_+_169659099
|
0.858
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr3_-_46249787
|
0.776
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr1_+_182569427
|
0.756
|
|
|
|
|
chr5_+_71502700
|
0.730
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr20_+_2276612
|
0.720
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr17_+_43299277
|
0.719
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr3_-_164914448
|
0.709
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr3_+_119013216
|
0.708
|
NM_020754
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr2_-_158182154
|
0.700
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr6_+_33239798
|
0.688
|
NM_022551
|
RPS18
|
ribosomal protein S18
|
|
chr17_-_43487747
|
0.650
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr9_+_108424737
|
0.635
|
NM_005421
|
TAL2
|
T-cell acute lymphocytic leukemia 2
|
|
chr17_+_45908992
|
0.635
|
NM_033413
|
LRRC46
|
leucine rich repeat containing 46
|
|
chr10_+_114154522
|
0.608
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr19_-_16606938
|
0.607
|
NM_145046
|
CALR3
|
calreticulin 3
|
|
chr12_-_123214882
|
0.607
|
NM_032554
|
HCAR1
|
hydroxycarboxylic acid receptor 1
|
|
chr7_-_107880492
|
0.604
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr3_-_12200339
|
0.580
|
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr4_+_87857394
|
0.578
|
|
|
|
|
chr6_+_33239855
|
0.559
|
|
RPS18
|
ribosomal protein S18
|
|
chr7_+_100770378
|
0.544
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr12_+_109551041
|
0.536
|
|
|
|
|
chr15_+_41136622
|
0.528
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr6_+_33589045
|
0.520
|
NM_002224
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3
|
|
chr22_-_37640187
|
0.510
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr4_-_153601085
|
0.500
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr13_+_44947977
|
0.486
|
NM_001010897
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2
|
|
chr9_+_2621740
|
0.485
|
NM_001018056
NM_003383
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr4_+_84457390
|
0.482
|
NM_032717
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr7_+_140103842
|
0.482
|
NM_001008749
|
RAB19
|
RAB19, member RAS oncogene family
|
|
chr12_+_52306112
|
0.479
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr11_+_111789600
|
0.477
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr11_+_308106
|
0.471
|
NM_006435
|
IFITM2
|
interferon induced transmembrane protein 2 (1-8D)
|
|
chr1_+_171810611
|
0.469
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chrX_-_154688260
|
0.469
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated 2
coagulation factor VIII-associated 3
coagulation factor VIII-associated 1
|
|
chr12_-_53600999
|
0.464
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr2_-_238499713
|
0.458
|
NM_022449
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr6_-_154677868
|
0.448
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr5_-_147763345
|
0.443
|
|
|
|
|
chr2_-_175499275
|
0.439
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr8_-_6420454
|
0.439
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr12_-_58159386
|
0.438
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr16_-_279110
|
0.435
|
|
|
|
|
chr12_-_57871631
|
0.434
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr4_+_84457256
|
0.431
|
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr1_+_206730893
|
0.421
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr12_-_51785192
|
0.415
|
NM_007210
|
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6)
|
|
chr7_+_18535351
|
0.415
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chrX_-_15402469
|
0.407
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr1_+_171283372
|
0.401
|
NM_002022
|
FMO4
|
flavin containing monooxygenase 4
|
|
chr10_+_75545296
|
0.401
|
|
KIAA0913
|
KIAA0913
|
|
chr1_-_111743261
|
0.398
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr14_+_77924367
|
0.393
|
NM_012111
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast)
|
|
chr9_+_75766777
|
0.392
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr14_-_75530716
|
0.389
|
NM_001107
NM_203488
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type
|
|
chr15_+_85359910
|
0.388
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr17_+_16318848
|
0.386
|
NM_016113
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr17_-_62097937
|
0.385
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr1_+_206730591
|
0.383
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr14_-_57960432
|
0.383
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr1_+_168545710
|
0.381
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr17_+_57915192
|
0.379
|
|
|
|
|
chr11_-_6790187
|
0.378
|
NM_001004490
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2
|
|
chrX_+_70315637
|
0.376
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr8_+_82192717
|
0.374
|
NM_001444
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated)
|
|
chr12_+_21679190
|
0.374
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr2_-_161056762
|
0.373
|
|
ITGB6
|
integrin, beta 6
|
|
chr7_+_18548899
|
0.373
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr5_-_172083569
|
0.372
|
|
|
|
|
chr12_-_123590045
|
0.372
|
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr11_+_7506599
|
0.371
|
NM_198474
|
OLFML1
|
olfactomedin-like 1
|
|
chr14_+_77924415
|
0.367
|
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast)
|
|
chr17_-_39093671
|
0.367
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr19_-_49258646
|
0.366
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr1_+_206730497
|
0.363
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr7_+_18548963
|
0.361
|
|
HDAC9
|
histone deacetylase 9
|
|
chr11_-_118213321
|
0.359
|
NM_000732
NM_001040651
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex)
|
|
chr17_+_48351787
|
0.357
|
NM_153229
|
TMEM92
|
transmembrane protein 92
|
|
chr2_+_135011829
|
0.354
|
NM_002410
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase
|
|
chr4_+_39529551
|
0.353
|
|
|
|
|
chr12_+_6881669
|
0.352
|
NM_002286
|
LAG3
|
lymphocyte-activation gene 3
|
|
chr5_-_75919051
|
0.351
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr1_-_168513201
|
0.350
|
NM_003175
|
XCL1
XCL2
|
chemokine (C motif) ligand 1
chemokine (C motif) ligand 2
|
|
chr6_-_87804773
|
0.341
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr8_+_22437983
|
0.341
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr1_-_27286875
|
0.340
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chr1_+_206730456
|
0.339
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr16_+_50300461
|
0.339
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr15_+_41136178
|
0.338
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr2_-_238499317
|
0.338
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr7_-_143892649
|
0.338
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr6_+_12290528
|
0.334
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr7_+_37960418
|
0.329
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr9_+_75766794
|
0.329
|
|
ANXA1
|
annexin A1
|
|
chr5_-_176037091
|
0.329
|
NM_052899
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr2_-_161056573
|
0.327
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr1_-_202679415
|
0.326
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr16_+_4845265
|
0.326
|
NM_001253793
NM_001253794
|
LOC440335
|
uncharacterized LOC440335
|
|
chr19_+_57742376
|
0.325
|
NM_001015879
NM_003160
NM_001015878
|
AURKC
|
aurora kinase C
|
|
chr15_+_59908908
|
0.325
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr9_+_103115342
|
0.322
|
|
|
|
|
chr6_-_111888520
|
0.320
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr9_+_103204187
|
0.319
|
NM_001198812
|
C9orf30-TMEFF1
C9orf30
|
C9orf30-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chr14_+_77227783
|
0.318
|
|
|
|
|
chr20_-_35201677
|
0.318
|
|
|
|
|
chr10_+_695887
|
0.316
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr6_+_54173202
|
0.316
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr6_+_139349877
|
0.315
|
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr11_-_62389571
|
0.314
|
NM_012200
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr4_+_186317837
|
0.314
|
NM_181726
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr17_-_62084282
|
0.312
|
NM_000873
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr6_+_139349818
|
0.311
|
NM_021243
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr10_+_43633880
|
0.310
|
NM_018590
|
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2
|
|
chr17_-_7832752
|
0.310
|
NM_004732
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3
|
|
chr9_-_77703040
|
0.309
|
NM_001127603
NM_017881
|
C9orf95
|
chromosome 9 open reading frame 95
|
|
chr11_-_104034685
|
0.309
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr5_-_43412417
|
0.308
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr11_-_62389376
|
0.308
|
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr2_+_177016112
|
0.308
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr19_-_13261088
|
0.307
|
|
STX10
|
syntaxin 10
|
|
chr3_-_185270258
|
0.306
|
NM_139248
|
LIPH
|
lipase, member H
|
|
chr1_-_93811322
|
0.303
|
|
LOC100131564
|
uncharacterized LOC100131564
|
|
chr8_-_8175860
|
0.302
|
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr17_-_9479139
|
0.300
|
NM_004853
|
STX8
|
syntaxin 8
|
|
chr19_+_49838673
|
0.299
|
NM_001040031
NM_001774
|
CD37
|
CD37 molecule
|
|
chr10_-_118680978
|
0.299
|
|
KIAA1598
|
KIAA1598
|
|
chr6_+_31582978
|
0.297
|
NM_004847
NM_001623
|
AIF1
|
allograft inflammatory factor 1
|
|
chr2_+_37458773
|
0.296
|
NM_001083946
NM_144736
|
C2orf56
|
chromosome 2 open reading frame 56
|
|
chr17_+_16319087
|
0.293
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr12_+_104682495
|
0.292
|
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr4_-_84255895
|
0.290
|
NM_001199830
|
HPSE
|
heparanase
|
|
chr7_-_100061893
|
0.288
|
NM_001004323
|
C7orf61
|
chromosome 7 open reading frame 61
|
|
chr18_+_3594056
|
0.288
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr1_-_147931877
|
0.286
|
|
FLJ39739
|
uncharacterized FLJ39739
|
|
chr1_+_154300275
|
0.286
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr3_-_13057130
|
0.286
|
|
|
|
|
chr7_+_76178604
|
0.284
|
|
LOC100133091
|
uncharacterized LOC100133091
|
|
chr20_+_19870209
|
0.282
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr7_+_37960162
|
0.282
|
NM_001242946
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr19_-_51014344
|
0.282
|
NM_138334
|
JOSD2
|
Josephin domain containing 2
|
|
chr11_+_72980938
|
0.281
|
NM_176798
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr11_+_64008412
|
0.280
|
NM_004470
|
FKBP2
|
FK506 binding protein 2, 13kDa
|
|
chr8_-_119634116
|
0.279
|
|
SAMD12
|
sterile alpha motif domain containing 12
|
|
chr1_-_32687830
|
0.276
|
NM_019118
|
TMEM234
|
transmembrane protein 234
|
|
chr8_+_9953061
|
0.275
|
NM_001135671
NM_001199729
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr11_+_63974190
|
0.273
|
|
FERMT3
|
fermitin family member 3
|
|
chr19_+_36203829
|
0.272
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr6_-_41673677
|
0.272
|
|
TFEB
|
transcription factor EB
|
|
chrX_-_47489243
|
0.271
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr18_+_3453771
|
0.271
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_+_40506423
|
0.269
|
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|
|
chr2_+_121103670
|
0.268
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr11_-_64764388
|
0.268
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chr3_-_139258531
|
0.265
|
NM_001130992
NM_001130993
NM_002899
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr19_+_47813074
|
0.265
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr6_-_43595079
|
0.265
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr20_+_388941
|
0.264
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr1_-_201368617
|
0.264
|
NM_005558
|
LAD1
|
ladinin 1
|
|
chr5_+_145718586
|
0.263
|
NM_002700
|
POU4F3
|
POU class 4 homeobox 3
|
|
chr2_+_148778573
|
0.263
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr11_-_111944757
|
0.259
|
NM_001082619
NM_138789
|
PIH1D2
|
PIH1 domain containing 2
|
|
chr9_+_3526047
|
0.258
|
|
|
|
|
chr21_+_26934440
|
0.255
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr5_-_172756505
|
0.255
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr1_+_198608216
|
0.255
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr7_+_141463896
|
0.254
|
NM_016943
|
TAS2R3
|
taste receptor, type 2, member 3
|
|
chr8_+_99076749
|
0.253
|
NM_001170806
NM_173549
|
C8orf47
|
chromosome 8 open reading frame 47
|
|
chr2_+_28615695
|
0.248
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr14_-_24036939
|
0.247
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr1_-_111743297
|
0.247
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr10_-_15902455
|
0.246
|
NM_024948
|
FAM188A
|
family with sequence similarity 188, member A
|
|
chr2_-_175462428
|
0.246
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr4_+_110834039
|
0.245
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr11_-_65638672
|
0.245
|
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr2_+_65663812
|
0.245
|
|
FLJ16124
|
FLJ16124 protein
|
|
chr14_-_24037266
|
0.243
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr13_+_46626949
|
0.240
|
|
LOC100509894
|
uncharacterized LOC100509894
|
|
chr17_+_48348766
|
0.240
|
NM_001168215
|
TMEM92
|
transmembrane protein 92
|
|
chr19_+_17530841
|
0.240
|
|
FAM125A
|
family with sequence similarity 125, member A
|
|
chr9_-_130617009
|
0.239
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr1_+_40506406
|
0.239
|
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|
|
chr1_-_32687939
|
0.239
|
|
TMEM234
|
transmembrane protein 234
|
|
chr5_+_125758818
|
0.238
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr6_-_43595046
|
0.238
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr14_+_45366426
|
0.237
|
NM_001017923
|
C14orf28
|
chromosome 14 open reading frame 28
|
|
chrX_+_12885190
|
0.237
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr5_-_171615095
|
0.237
|
|
STK10
|
serine/threonine kinase 10
|
|
chr2_-_133539627
|
0.236
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr19_+_50879681
|
0.236
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr16_-_30107492
|
0.234
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr12_-_62997207
|
0.234
|
NM_175895
|
C12orf61
|
chromosome 12 open reading frame 61
|
|
chr12_-_58135853
|
0.234
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr5_+_59784004
|
0.234
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr2_+_241544819
|
0.234
|
NM_001195381
NM_001195382
|
GPR35
|
G protein-coupled receptor 35
|