|
chr6_+_26045611
|
2.250
|
NM_003531
|
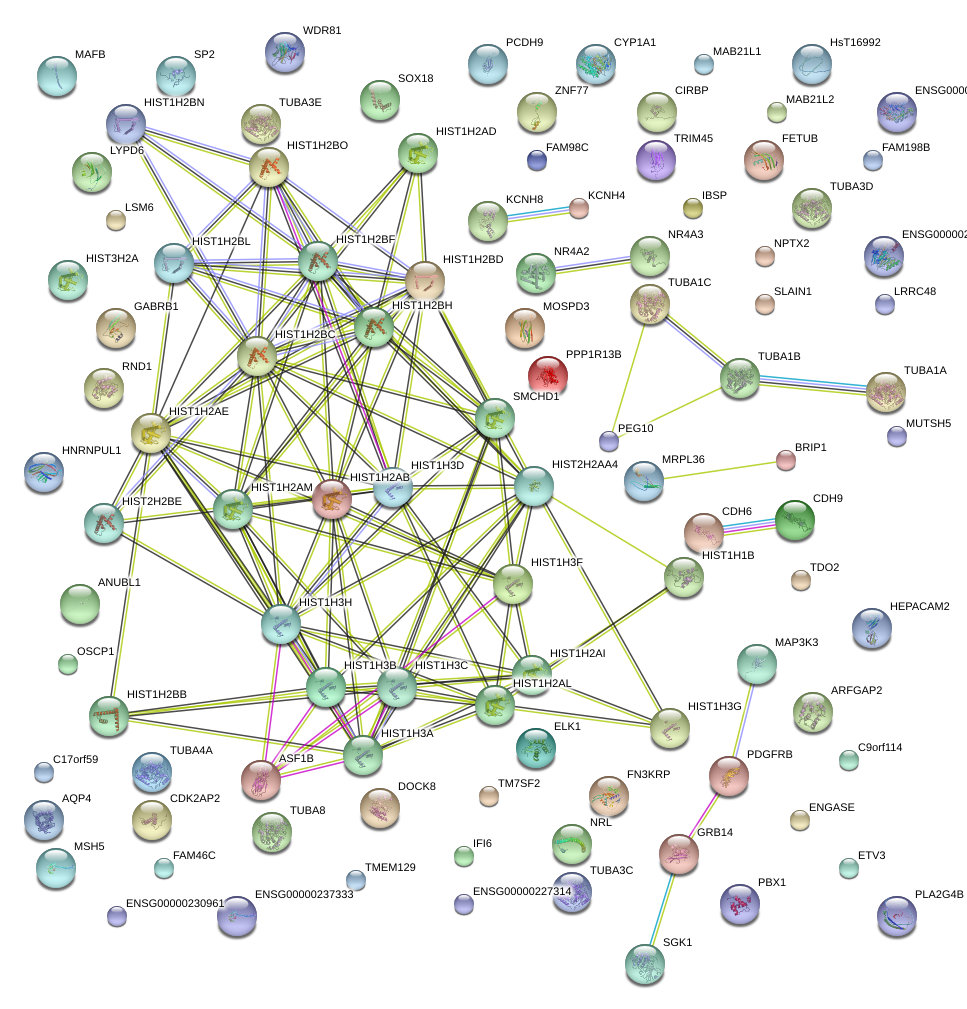
HIST1H3C
|
histone cluster 1, H3c
|
|
chr1_+_151763174
|
1.748
|
|
|
|
|
chr6_+_27775898
|
1.567
|
NM_003509
|
HIST1H2AI
HIST1H2AM
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H2am
histone cluster 1, H3f
|
|
chr6_+_26199712
|
1.347
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr5_-_27038650
|
1.290
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr13_-_67804432
|
1.275
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr6_+_26217145
|
1.226
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr4_+_156824846
|
1.180
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr6_-_27775683
|
1.116
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr5_+_31193852
|
1.090
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr6_+_27833033
|
1.088
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr1_-_36915969
|
0.976
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr4_+_88720701
|
0.908
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr4_+_47033294
|
0.889
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr11_-_67275549
|
0.883
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr1_-_117664349
|
0.844
|
NM_001145635
NM_025188
|
TRIM45
|
tripartite motif containing 45
|
|
chr19_-_2944894
|
0.823
|
NM_021217
|
ZNF77
|
zinc finger protein 77
|
|
chr3_-_169482847
|
0.816
|
|
TERC
|
telomerase RNA component
|
|
chr4_+_151503076
|
0.808
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr19_+_20959109
|
0.802
|
|
|
|
|
chr17_+_20771745
|
0.783
|
|
|
|
|
chr2_-_157189040
|
0.776
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr19_+_38893766
|
0.759
|
NM_174905
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr6_-_26032287
|
0.747
|
NM_003537
|
HIST1H3B
|
histone cluster 1, H3b
|
|
chr7_-_92855780
|
0.740
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr6_-_27835308
|
0.696
|
NM_005322
|
HIST1H1B
|
histone cluster 1, H1b
|
|
chr6_-_26043884
|
0.673
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr4_-_146859960
|
0.665
|
|
|
|
|
chr18_-_24445652
|
0.641
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr19_-_14247400
|
0.635
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr19_-_14247267
|
0.625
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr5_+_31193761
|
0.624
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr17_+_17876126
|
0.608
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr7_+_100209724
|
0.595
|
NM_001040097
|
MOSPD3
|
motile sperm domain containing 3
|
|
chr6_+_26251826
|
0.584
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr7_+_94285676
|
0.582
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr9_+_214816
|
0.577
|
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr7_+_94285736
|
0.575
|
|
PEG10
|
paternally expressed 10
|
|
chr17_+_80674579
|
0.570
|
NM_024619
|
FN3KRP
|
fructosamine 3 kinase related protein
|
|
chr15_+_42131010
|
0.540
|
NM_001114633
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic)
|
|
chr17_-_8093346
|
0.535
|
|
C17orf59
|
chromosome 17 open reading frame 59
|
|
chr17_+_45973515
|
0.528
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr5_-_149535420
|
0.519
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr5_-_149535138
|
0.518
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr11_+_10830752
|
0.517
|
|
|
|
|
chr2_-_73460326
|
0.511
|
NM_032319
|
PRADC1
|
protease-associated domain containing 1
|
|
chr11_-_47198379
|
0.501
|
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr7_+_94285620
|
0.500
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr12_-_49259549
|
0.498
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr3_-_190580464
|
0.495
|
NM_001146686
|
GMNC
|
geminin coiled-coil domain containing
|
|
chr15_-_75017710
|
0.493
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr1_-_27998658
|
0.490
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr20_-_39317875
|
0.482
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr7_+_98246595
|
0.481
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr3_+_186358148
|
0.480
|
NM_014375
|
FETUB
|
fetuin B
|
|
chr6_-_134498973
|
0.478
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_-_27998719
|
0.477
|
NM_002038
NM_022872
NM_022873
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr6_-_26033795
|
0.472
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr3_+_50242688
|
0.471
|
NM_006841
|
SLC38A3
|
solute carrier family 38, member 3
|
|
chr13_+_78315294
|
0.466
|
NM_001242869
NM_001242870
NM_001242871
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr1_+_43312288
|
0.465
|
|
|
|
|
chr11_-_47198420
|
0.461
|
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr19_+_41770057
|
0.457
|
NM_007040
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr5_-_149535309
|
0.457
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr4_+_147096834
|
0.455
|
NM_007080
|
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr17_-_59940669
|
0.445
|
NM_032043
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1
|
|
chr1_+_164528914
|
0.443
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_+_118148668
|
0.442
|
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr6_-_26197477
|
0.436
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr17_+_61699786
|
0.434
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr2_+_132233579
|
0.428
|
NM_080386
|
TUBA3D
|
tubulin, alpha 3d
|
|
chr19_+_1269244
|
0.426
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr2_-_165424993
|
0.425
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr9_-_131592072
|
0.422
|
|
C9orf114
|
chromosome 9 open reading frame 114
|
|
chr17_+_61699783
|
0.422
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr6_-_26271588
|
0.421
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr19_+_41770273
|
0.421
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr2_+_150187028
|
0.421
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr14_-_104313730
|
0.418
|
NM_015316
|
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B
|
|
chr4_-_159094163
|
0.417
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr11_+_64879294
|
0.416
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr10_-_46168156
|
0.416
|
NM_001128324
NM_174890
|
ZFAND4
|
zinc finger, AN1-type domain 4
|
|
chr17_+_1619816
|
0.414
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr4_-_775586
|
0.414
|
|
LOC100129917
|
uncharacterized LOC100129917
|
|
chr20_-_62680880
|
0.412
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr6_+_31707724
|
0.410
|
NM_002441
NM_025259
NM_172165
NM_172166
|
MSH5-C6orf26
MSH5
|
MSH5-C6orf26 readthrough (non-protein coding)
mutS homolog 5 (E. coli)
|
|
chr14_-_24584125
|
0.408
|
|
NRL
|
neural retina leucine zipper
|
|
chr4_-_1722776
|
0.406
|
|
TMEM129
|
transmembrane protein 129
|
|
chr17_+_77071002
|
0.406
|
NM_001042573
|
ENGASE
|
endo-beta-N-acetylglucosaminidase
|
|
chrX_-_47509864
|
0.394
|
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr4_+_4861371
|
0.393
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr10_-_93392799
|
0.389
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr4_-_1722978
|
0.388
|
NM_001127266
NM_138385
|
TMEM129
|
transmembrane protein 129
|
|
chr5_-_73937248
|
0.387
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr4_-_100065372
|
0.386
|
NM_000670
|
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide
|
|
chr16_-_67965754
|
0.385
|
NM_001907
|
CTRL
|
chymotrypsin-like
|
|
chr19_+_36505409
|
0.382
|
|
|
|
|
chr10_-_101690655
|
0.381
|
|
DNMBP
|
dynamin binding protein
|
|
chr2_-_45236521
|
0.380
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr6_-_26199439
|
0.380
|
NM_021065
NM_003530
|
HIST1H2AD
HIST1H3D
|
histone cluster 1, H2ad
histone cluster 1, H3d
|
|
chr1_-_149859465
|
0.378
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr17_-_76183074
|
0.378
|
NM_003258
|
TK1
|
thymidine kinase 1, soluble
|
|
chr6_-_26216871
|
0.378
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr16_-_79634610
|
0.378
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr5_-_137090049
|
0.377
|
|
|
|
|
chr19_+_14640363
|
0.376
|
NM_138501
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr17_-_35293920
|
0.375
|
|
|
|
|
chr11_+_34460533
|
0.372
|
|
CAT
|
catalase
|
|
chr6_-_27100528
|
0.368
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr6_-_30685457
|
0.368
|
NM_014641
|
MDC1
|
mediator of DNA-damage checkpoint 1
|
|
chr11_-_19263144
|
0.367
|
|
E2F8
|
E2F transcription factor 8
|
|
chrX_+_13671224
|
0.366
|
NM_152634
|
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing
|
|
chr19_+_14640406
|
0.365
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr19_-_55972935
|
0.363
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr6_+_26156552
|
0.362
|
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr1_+_118148506
|
0.362
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr22_+_19467467
|
0.362
|
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr5_-_35230548
|
0.361
|
|
PRLR
|
prolactin receptor
|
|
chr1_+_167298382
|
0.360
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_+_111408648
|
0.359
|
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr16_-_30596808
|
0.359
|
NM_152458
|
ZNF785
|
zinc finger protein 785
|
|
chr6_-_153304093
|
0.359
|
NM_012177
|
FBXO5
|
F-box protein 5
|
|
chr17_-_1928177
|
0.359
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr16_+_72088507
|
0.358
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr6_-_135271206
|
0.356
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr22_+_19467413
|
0.356
|
NM_001178010
NM_001178011
NM_003504
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr19_+_14640413
|
0.356
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chrX_+_52926358
|
0.354
|
|
FAM156B
|
family with sequence similarity 156, member B
|
|
chr6_+_26156558
|
0.354
|
NM_005321
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr10_-_21807120
|
0.353
|
|
|
|
|
chr14_+_53019865
|
0.352
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr19_-_2042022
|
0.352
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr19_+_2841432
|
0.348
|
NM_001172775
NM_152791
|
ZNF555
|
zinc finger protein 555
|
|
chr19_+_39616384
|
0.344
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr2_-_86564632
|
0.343
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr8_+_27631902
|
0.342
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr14_-_105487365
|
0.341
|
|
CDCA4
|
cell division cycle associated 4
|
|
chr17_-_8093492
|
0.339
|
NM_017622
|
C17orf59
|
chromosome 17 open reading frame 59
|
|
chr9_+_34989637
|
0.338
|
NM_001135004
NM_012266
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr8_+_37553269
|
0.336
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr12_+_117176085
|
0.336
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr16_+_8806825
|
0.336
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr2_-_180427314
|
0.335
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr6_-_27860927
|
0.334
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chr17_-_1619473
|
0.334
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chr1_+_179051218
|
0.333
|
|
TOR3A
|
torsin family 3, member A
|
|
chr10_-_131909061
|
0.332
|
|
LOC387723
|
uncharacterized LOC387723
|
|
chr10_-_93392744
|
0.331
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr14_-_105487421
|
0.330
|
NM_017955
NM_145701
|
CDCA4
|
cell division cycle associated 4
|
|
chr11_+_64052282
|
0.330
|
NM_001177358
NM_001170881
|
GPR137
|
G protein-coupled receptor 137
|
|
chr7_-_148725731
|
0.330
|
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr2_+_111878450
|
0.330
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr17_+_61699715
|
0.329
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr5_-_137090033
|
0.329
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr11_-_64052164
|
0.328
|
NM_004322
NM_032989
|
BAD
|
BCL2-associated agonist of cell death
|
|
chr3_+_181430075
|
0.325
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr8_+_118532953
|
0.323
|
NM_080651
|
MED30
|
mediator complex subunit 30
|
|
chr11_-_19262424
|
0.322
|
NM_024680
|
E2F8
|
E2F transcription factor 8
|
|
chr9_+_132083294
|
0.322
|
NM_001012715
|
C9orf106
|
chromosome 9 open reading frame 106
|
|
chr17_+_43225834
|
0.320
|
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr17_+_76183397
|
0.320
|
NM_001010982
NM_001145526
|
AFMID
|
arylformamidase
|
|
chr21_+_44313359
|
0.317
|
NM_001001503
NM_021075
|
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa
|
|
chr11_-_129872679
|
0.316
|
|
PRDM10
|
PR domain containing 10
|
|
chr14_-_105487413
|
0.316
|
|
CDCA4
|
cell division cycle associated 4
|
|
chr1_+_164528586
|
0.316
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr5_+_36152142
|
0.316
|
NM_001243120
NM_005983
NM_032637
|
SKP2
|
S-phase kinase-associated protein 2 (p45)
|
|
chr10_-_93392857
|
0.315
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr5_-_150138081
|
0.313
|
NM_001135644
|
DCTN4
|
dynactin 4 (p62)
|
|
chr2_+_71127692
|
0.312
|
|
VAX2
|
ventral anterior homeobox 2
|
|
chr17_+_7531275
|
0.312
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr17_+_43225921
|
0.311
|
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr6_+_111408780
|
0.308
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr5_+_36152221
|
0.308
|
|
SKP2
|
S-phase kinase-associated protein 2 (p45)
|
|
chr10_+_76586299
|
0.305
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr20_-_32307889
|
0.305
|
NM_007238
NM_183397
|
PXMP4
|
peroxisomal membrane protein 4, 24kDa
|
|
chr8_+_25316512
|
0.304
|
NM_152562
|
CDCA2
|
cell division cycle associated 2
|
|
chr17_-_43025027
|
0.304
|
NM_001080443
|
KIF18B
|
kinesin family member 18B
|
|
chr17_+_45727558
|
0.302
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr1_-_27998700
|
0.302
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr16_-_30621628
|
0.300
|
NM_138447
|
ZNF689
|
zinc finger protein 689
|
|
chr14_+_52456322
|
0.299
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr6_+_26183957
|
0.298
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr17_-_58212896
|
0.293
|
|
|
|
|
chr14_+_29236171
|
0.292
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr6_-_27858569
|
0.292
|
NM_003535
|
HIST1H3J
|
histone cluster 1, H3j
|
|
chr19_+_38893804
|
0.291
|
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr3_+_50273635
|
0.288
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr8_+_118532970
|
0.287
|
|
MED30
|
mediator complex subunit 30
|
|
chrX_-_15353634
|
0.287
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr15_-_72612247
|
0.286
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr5_+_140529619
|
0.284
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr10_+_102222797
|
0.283
|
NM_003393
|
WNT8B
|
wingless-type MMTV integration site family, member 8B
|
|
chr14_-_74551159
|
0.283
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr16_+_2933195
|
0.282
|
NM_001142499
NM_001142500
NM_138439
|
FLYWCH2
|
FLYWCH family member 2
|
|
chr2_-_190044330
|
0.282
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_+_169492051
|
0.282
|
NM_001185119
|
MYNN
|
myoneurin
|
|
chr17_-_8198605
|
0.281
|
|
SLC25A35
|
solute carrier family 25, member 35
|
|
chr16_-_74455239
|
0.280
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr4_-_174255454
|
0.278
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr11_+_64051810
|
0.278
|
NM_001170726
|
GPR137
|
G protein-coupled receptor 137
|